Gene
KWMTBOMO11088
Pre Gene Modal
BGIBMGA011048
Annotation
reverse_transcriptase_[Operophtera_brumata]
Location in the cell
Nuclear Reliability : 3.783
Sequence
CDS
ATGGTAAATACCCGCAGTACCGCCAACAAACAAAGAAAAAACGACGATGAGCATGACGCAAAAAGTTCTTGTGCCGCGAGTGCAGTGAAGTCGGAACGAACCGAAAAACAAAATCCGCCTTCACTAGCATCACGTCAGTCAGTCAGGTCCAAGAAAATGTCATCATCAAAATCAATACAAATCCAGAAAAAGAAACTGGAACTAGAAGCAGCTCAGGCGAAGGCGCGTATTCAAATGGAACTCATTGATCAAAAGTTAGCGGCGGATCTAGCAGAACTCGAAGAAGAATACAGTCCACCATCAGAAATATCGGAGAAAAACAACGGCGGTGATATAGAAAAATGGTTAGAGCGTAGTCAACAGGAACTGCAGCAGGCCACTAAGCTCAACAGTGATAAACACAAGTCACACTCAAGCCCGGCCGACAACGACGCAGTTCAACAACTCACGAGCGCATTCAAAGAATTTATGACGTCCTCCGCCATCCAGCAAAATACTAAGCTGCTTAGTCGTCTCAGCACTCCAAAAGAACTCCCTATCTTCAGCGGAGACCCAAAAGAATGGCTTTATTTCGAGCAGGCCTACCTGGAATCGACTGAAGTCTGCAACTTCAGCGATAAAGAAAACCATTGGCGGTTACGAAAGTGTTTACGAGGTACCGCACAAGAAGCCGTCGCCGGCCTACTTATCAGTGGTGCTTCTCCTGCAGAAATAATTTCCACTTTAAAACTACAGTTCGGAAACGCTGACGTCATTCTATCTCGCATCGTGGTAGACGTAAAGAAGATGAATCCACTGCCTACGGACTACCACAAAGAAATAGTACCCTTTTCAGGAAAAGTGAAAAATTACGTGGCCGCTGTACGAGCCCTTGGTCGCCAAGAGTACCTGCAAGGCATGGAAGTGGCTAACATTATCTTATGCAAGCTACCCACGACCCTGCTCACCAAGTGGACAGATTATAGTTATCCGCTTATCACAGCTGAATCGCCGAAACCTAGACTAGAGGTGCTGTCAGATTTCCTCACCGAAGAAGCTGTAAAAGTATCAACCTGTAGTGCTACTTTATTGCAATCTAGGACGGACAATTTCAAACGTAAACTTTATGATAAAGTGAACAATATACAAACAATTTTAGTGCAAGACGGACAAAGTGTACAAAAATGTGGTTATTGTCGTACCGGTAGCCATAAATTAACCGAGTGTAAAAAATTTAAAAAATCACTAAGAAAGGACAGATGGTTCCATGTGAAGCGTTACGGCATTTGTTATAAGTGTTTATTAGCGCATCATCGCAGAGAAACGTGCTCCGCTCCAAGCTGTGACGTAAGCAATTGTGGACAATCGCATCACCGCTTACTGCATTATGTGCCGAGTCGAAACGACACTCAATCTGTATCAGCAGTACAATCTGAAACGCCACCTGAAACTGAAGTTCAGTCTGAAATAGTAACAAATATTAATGTCGGCTGTTCCGTAATGTTAAAAATTGTGCCTATAAATATTCACGGACCGAACGGAATTGTTAGCACGACCGCCTTGCTCGACGATGGCTCGACTGTGTCGTTGATTAGCGCTCCGCTGGCATTACAAGTGGGACTGCGCGGCCGCCACGAACGTCTTCGTGTGCGCGGCGCGTGGAACGACACTGAATTAGTGTGCGATGCGCAGCTAGCAAACGTGACACTATGTAGTAAGTGTAGTACCGATAACAAGATGTTTACAATCGATGCGCGGATTGTCGAAGAGTTAAACCTACCACAAAGTTTGCCTACAGTAAATGTTAAAAAATATTTGCACCTAAATGACATCAGCCATAACTTATGCACCACAACACAACTGCATCCGCAAGTTTTAATTGGACAAGATAATTGTCACCTCCTCTTACCTCTGGAAATGAGAGTGGGTAAGCCCGATGAACCCTGTGCCACCCGGACTCCTCTGGGTTGGTGCCTCCATGGAAGAGTACCTAATGCTTACCTACCTAATTCTGCTGCACGTCACGCTTCATTGTTTGTCTCCAGTGACGTCATAGAATCTGAATGTACTTTGCGAGAGATCCACGAGGAGGTGCGAAGATCGTTCTCAATCGAGTCCATGGGCGTGTCTGGCAAACCAAGACAGAACTCAGAAGATTTACGCGCTGTTGAAATTTTAGAACGAACCTCCACTCTCGTCAATGGTCAATGGCATGTTGGCTTACCTTGGCGGGATGAGACTTGTACGATGCCGGACTCGTACCTAACAGCTCTCCGTAGACTAAAAGGAATCGAACAAAAATTAAAAAATAATAAAGAATATGCAAAAAGATACGAAGAACGAATACGACATTCATTTGAAAATGATTTCGCAAAAAAACTGATCAGCTCAAATGTTACACCACTGAAAACTTGGTATTTACCTCATTTTGGTGTCGATAACCCGAACAAAAAGAAACTGCGCTTAGTGTTTGACGCCGCGGCCAAAACAAATGGTTTGTCCCTGAATGATTACTTATTGAAAGGGCCCGACCTACTAATGTCATTATTCGCCATCATGTTGCGATTTAGAGAAAATCGAATAGCCGTTTCTGGAGATATTAAAGATATGTTTCTCAAAGTGAAAATTCGACCAAAAAACCAGGATGCCTTCCGTTTTTTATATAGAGAAGAACCGGAGGGAAATATAAACACTTACGTTATGACGTCATTAATTTTTGGTGCTAATTGTTCGCCATTTATAGCTCAATTTGTAAAAAATAAAAACGCTCAGCGGTATGAGTCATCAATGCCCGCTGCTGCTTCCGCCATATATAGGAGTCACTATATGGACTATATCCACTATATGGATGATTACATAACCGAAGCGGTATTCCAGAAATGGTTGAAATGGATTTATCTTCTGGAATCATTAAAAGACGTGAGTTTGCCGAGGTACTATCCGGCTGCTACCATACGAGAGAATGAGACGAATAATGAAACCCTTCCGCTGTCATCCTCACACAACGCGATTTCTAGGACTACGGCTACGAATAATGCTACGAGCGTAGCACACGCCGAATCCGCTACGAATACTGCATCGCACAGCTACAATAATAATTGTGGAGACGATTACAGTGGAGTTTTAACTCGAACTGGATATAGAAACTTACAGATGCATATATTTTGTGATGCGTCTAACAAAGCATACTGCGCCGTCGCGTACTGGAGATGGATTGACGACGAGTTTAACATACCCGTAGCATTCATTGCGAGCAAATCTCGAGTTGCAGGAAACAAACCTATTACCATCCCACGTTTGGAACTTCAAGCCGCCGTCTTAGCGGCAAGACTAGCGGACTCAGTCACTAGAGAACATATTATAAAGCCTGAGCAACGAATATTTTGGAGCGACTCGACGACAGTGCTGCAATGGATAAGAAATGACGCTCGCGACTATAAAGTTTATGTGGCAAATCGTTTGGGCGAAATTGATGAGTTAACAAAGATAAAAGAATGGCGTTATGTTCCCACTAAATTAAACGTCGCTGATATTGCAACGAGAGAAAACTATAAATTCCCGGCCCTCCAAGATGAATGGTTGTATGGACCTACCTTCTTACGCCACGAAGCAAACGAATGGCCTCACGACTTCATAAGCGACAGAAAAGAAACACTGCCAGAATGTGTTAATTTGATAAAAGTAGAAGTACAGGAAATTAACTTACCTGTACCTGATGCTACAAGATTTAGTTCTTGGCTTCGCTTGCTTCGATCCACTTCATTAGTACTCAAATTTATAAACAAATGCAAAAAACTTACCTTTGAGTTCGACGATGCTATGGAGAAAGCTGAAATATTGTTACTGAAAAAATCACAAGCTGAATCGTATAAAAAAGAGTTAACTGAAATAAAAAACAGTGGATGTGTATCAAAAACAAGTAATATACTTACCTTGTCCCCTTACTTAGACGAGTATGGAGTTAAGAGTCGGTGGCCGCATCGGTGCTGCTACTGGCATTCTACCTTAATCAAAAACCCTATTATACTAGACGGAACAAGCTACATTGCTAGATTGATCGTGAAATATTATCATTATTACTACGCACACGGAAATCAAGAAACAATAGTAAATGAAGTCAAACAAAGATACTGGATCACAAGACTGAGACCTACCGTTAAACATATAACATCTAAATGCTCATTTTGTAGAATAAGAAAATCCAAACCAGAAGTATAA
Protein
MVNTRSTANKQRKNDDEHDAKSSCAASAVKSERTEKQNPPSLASRQSVRSKKMSSSKSIQIQKKKLELEAAQAKARIQMELIDQKLAADLAELEEEYSPPSEISEKNNGGDIEKWLERSQQELQQATKLNSDKHKSHSSPADNDAVQQLTSAFKEFMTSSAIQQNTKLLSRLSTPKELPIFSGDPKEWLYFEQAYLESTEVCNFSDKENHWRLRKCLRGTAQEAVAGLLISGASPAEIISTLKLQFGNADVILSRIVVDVKKMNPLPTDYHKEIVPFSGKVKNYVAAVRALGRQEYLQGMEVANIILCKLPTTLLTKWTDYSYPLITAESPKPRLEVLSDFLTEEAVKVSTCSATLLQSRTDNFKRKLYDKVNNIQTILVQDGQSVQKCGYCRTGSHKLTECKKFKKSLRKDRWFHVKRYGICYKCLLAHHRRETCSAPSCDVSNCGQSHHRLLHYVPSRNDTQSVSAVQSETPPETEVQSEIVTNINVGCSVMLKIVPINIHGPNGIVSTTALLDDGSTVSLISAPLALQVGLRGRHERLRVRGAWNDTELVCDAQLANVTLCSKCSTDNKMFTIDARIVEELNLPQSLPTVNVKKYLHLNDISHNLCTTTQLHPQVLIGQDNCHLLLPLEMRVGKPDEPCATRTPLGWCLHGRVPNAYLPNSAARHASLFVSSDVIESECTLREIHEEVRRSFSIESMGVSGKPRQNSEDLRAVEILERTSTLVNGQWHVGLPWRDETCTMPDSYLTALRRLKGIEQKLKNNKEYAKRYEERIRHSFENDFAKKLISSNVTPLKTWYLPHFGVDNPNKKKLRLVFDAAAKTNGLSLNDYLLKGPDLLMSLFAIMLRFRENRIAVSGDIKDMFLKVKIRPKNQDAFRFLYREEPEGNINTYVMTSLIFGANCSPFIAQFVKNKNAQRYESSMPAAASAIYRSHYMDYIHYMDDYITEAVFQKWLKWIYLLESLKDVSLPRYYPAATIRENETNNETLPLSSSHNAISRTTATNNATSVAHAESATNTASHSYNNNCGDDYSGVLTRTGYRNLQMHIFCDASNKAYCAVAYWRWIDDEFNIPVAFIASKSRVAGNKPITIPRLELQAAVLAARLADSVTREHIIKPEQRIFWSDSTTVLQWIRNDARDYKVYVANRLGEIDELTKIKEWRYVPTKLNVADIATRENYKFPALQDEWLYGPTFLRHEANEWPHDFISDRKETLPECVNLIKVEVQEINLPVPDATRFSSWLRLLRSTSLVLKFINKCKKLTFEFDDAMEKAEILLLKKSQAESYKKELTEIKNSGCVSKTSNILTLSPYLDEYGVKSRWPHRCCYWHSTLIKNPIILDGTSYIARLIVKYYHYYYAHGNQETIVNEVKQRYWITRLRPTVKHITSKCSFCRIRKSKPEV
Summary
Uniprot
Pubmed
Proteomes
Pfam
Interpro
SUPFAM
SSF53098
SSF53098
Gene 3D
ProteinModelPortal
Ontologies
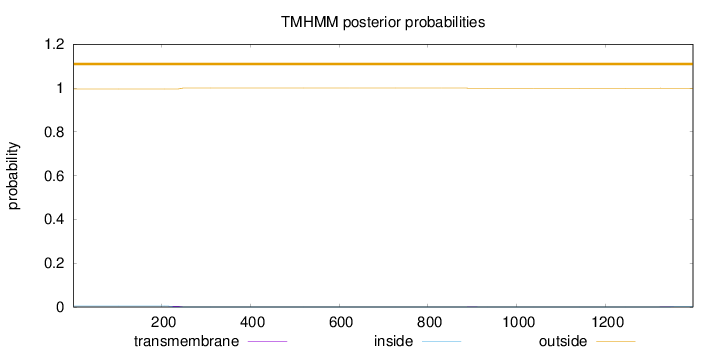
Topology
Length:
1398
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.10831
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00402
outside
1 - 1398
Population Genetic Test Statistics
Pi
217.132001
Theta
178.897903
Tajima's D
0.226054
CLR
1.371228
CSRT
0.430378481075946
Interpretation
Uncertain
