Pre Gene Modal
BGIBMGA008304
Annotation
PREDICTED:_RAC_serine/threonine-protein_kinase_[Bombyx_mori]
Full name
Non-specific serine/threonine protein kinase
+ More
RAC serine/threonine-protein kinase
RAC serine/threonine-protein kinase
Alternative Name
Akt
Protein kinase B
Protein kinase B
Location in the cell
Cytoplasmic Reliability : 2.236
Sequence
CDS
ATGGCGGCGCCCGGGATCATCGTGAAGGAAGGATGGCTGCAGAAACGTGGTGAACACATCCGGAATTGGCGAGATCGGTACTTCATATTGTTCGACACGGGGGACCTGGTGGGCTTCAAGACCCAGCCGGAGCGGAACAACTACAGGGACCCTCTCAACAAGTTCACCGTACGCGACTGCCAGATCATGGCTGTGGACAAACCTCGCCCCTACACTTTTACCATACGTGGCCTGCAGTGGACCACTGTTATTGAAAGGAACTTCTCAGTTGACAACGAAAAAGAAAGAGAAGAATGGGTGAAGGCCATCCGAGAAGTTGCCTCGCAACTGAGCACCGGAGGGCCCAGTTCAGCATCGATGTCCGATGCTGACGATCGCGACATGGCACAGCTCGGCACCAGCTTCCGTGACCCCCGCCGCATCACCTTAGAAAAATTCGAATTCGTGAAGGTGCTAGGCAAGGGTACGTTTGGGAAGGTGGTTCTGAGCCGTGAGAAGGGAACGGGAAAGCTGTACGCTATGAAGATATTGAAGAAACACCTCATTATACAGAAGGATGAGGTCGCGCACACCATCACCGAGAACCGAGTGCTCAAGAAGACCAAGCATCCATTTTTGACGGCTCTCCGCTATTCCTTCCAAACGGCAGACCGTGTCTGCTTCGTAATGGAATACGCTAATGGCGGTGAATTGTTCTTTCATTTGTCCCGCGAGCGTTCGTTCACCGAGGACCGCACCAGGTTCTACGGAGCCGAGATAGTATCTGCTCTCGGGTACCTCCACTCCGAGGGCATTATATATCGGGATTTGAAACTCGAGAATTTGCTGCTAGACAAAGACGGACACATCAAGATTGCAGACTTCGGACTATGCAAGGTGAATATAACGTATGGACGCACGACCAAGACGTTCTGTGGCACCCCCGAGTACCTCGCCCCTGAGGTCATAGAGGACTCCGATTATGGTCCCGCCGTAGATTGGTTAGTTGCTAACTAA
Protein
MAAPGIIVKEGWLQKRGEHIRNWRDRYFILFDTGDLVGFKTQPERNNYRDPLNKFTVRDCQIMAVDKPRPYTFTIRGLQWTTVIERNFSVDNEKEREEWVKAIREVASQLSTGGPSSASMSDADDRDMAQLGTSFRDPRRITLEKFEFVKVLGKGTFGKVVLSREKGTGKLYAMKILKKHLIIQKDEVAHTITENRVLKKTKHPFLTALRYSFQTADRVCFVMEYANGGELFFHLSRERSFTEDRTRFYGAEIVSALGYLHSEGIIYRDLKLENLLLDKDGHIKIADFGLCKVNITYGRTTKTFCGTPEYLAPEVIEDSDYGPAVDWLVAN
Summary
Description
Serine/threonine kinase involved in various developmental processes (PubMed:9601646, PubMed:10587646, PubMed:10962553, PubMed:11740943, PubMed:12172554, PubMed:11872800, PubMed:14525946, PubMed:12893776, PubMed:15466161, PubMed:15712201). During early embryogenesis, acts as a survival protein (PubMed:9601646, PubMed:10962553). During mid-embryogenesis, phosphorylates and activates trh, a transcription factor required for tracheal cell fate determination (PubMed:11740943). Also regulates tracheal cell migration (PubMed:11740943, PubMed:14525946). Later in development, acts downstream of PI3K and Pk61C/PDK1 in the insulin receptor transduction pathway which regulates cell growth and organ size, by phosphorylating and antagonizing FOXO transcription factor (PubMed:10587646, PubMed:10962553, PubMed:11752451, PubMed:12893776, PubMed:25329475, PubMed:29025897, PubMed:24603715). Controls follicle cell size during oogenesis (PubMed:15712201). May also stimulate cell growth by phosphorylating Gig/Tsc2 and inactivating the Tsc complex (PubMed:12172554, PubMed:15466161). Dephosphorylation of 'Ser-586' by Phlpp triggers apoptosis and suppression of tumor growth (PubMed:10962553).
Catalytic Activity
ATP + L-seryl-[protein] = ADP + H(+) + O-phospho-L-seryl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
Subunit
Interacts with trbl.
Similarity
Belongs to the protein kinase superfamily.
Belongs to the protein kinase superfamily. AGC Ser/Thr protein kinase family. RAC subfamily.
Belongs to the membrane-bound acyltransferase family.
Belongs to the protein kinase superfamily. AGC Ser/Thr protein kinase family. RAC subfamily.
Belongs to the membrane-bound acyltransferase family.
Keywords
Alternative initiation
Apoptosis
ATP-binding
Cell membrane
Complete proteome
Cytoplasm
Developmental protein
Growth regulation
Kinase
Membrane
Nucleotide-binding
Phosphoprotein
Reference proteome
Transferase
Feature
chain RAC serine/threonine-protein kinase
splice variant In isoform A.
splice variant In isoform A.
Uniprot
B0FC95
A0A2A4J8L6
G3LIS8
A0A385KNU3
A0A194QGX8
A0A194RMU6
+ More
S4PL92 A0A212EWQ2 H9JFK7 A0A0L7LP67 A0A087TFB1 E0VM17 A0A0K8UFB8 A0A034VRF0 A0A2H8TGM0 A0A0M3TCD7 A0A067QWC1 D2A1F0 U5ESE8 A0A2S2Q587 J9JPP1 A0A1Y1LC46 A0A2L2Y9W6 W8BSF8 A0A0T6AVT9 A0A131Y2G3 A0A147BUH3 T1P884 A0A182WNJ4 A0A1S3DA70 A0A1I8PVS7 A0A2M3YYS8 A0A2M3YYW6 A0A1B6GA14 V5H036 A0A182NY76 A0A2R5LBS4 Q7QK56 A0A3F2YSG6 A0A182XT68 A0A182VCH1 A0A182MDR8 Q8INB9-2 A0A0B4LIA3 B4QXQ5 B4HLD8 A0A293M499 B3P0Y2 H1ZY63 H1ZY64 Q8INB9 T1JL53 A0A0V1LT75 A0A0V0X785 A0A0V1PFC8 A0A1Y3EJB3 A0A0V0SNC6 A0A023GNP7 A0A1E1X356 A0A131YY67 A0A0V1C341 A0A224YYY4 L7MAG4 L7RA14 A0A1E1XRF4 A0A0V1N108 F5HR84 A0A0V1C2F7 A0A131XFF7 A0A0V1C2D6 A0A0V1FWK3 A0A0V1JFA2 A0A0V1AG58 A0A077YWW3 A0A0V0VUE6 A0A3Q0J4F5 A0A1D1VR66 A0A210Q3T4 A0A159ZKT0 A0A0V0UG04 A0A0V0UHG7 A0A0V1N1E2 A0A085NKI5 V9KNB7 A0A1S3JCH1 B5THM6 A0A0N5DJ40 A0A0S2GL00 W5K6L6 A0A2D0Q8N4 A0A3B4DV68 A0A0S7KZT5 E7EXT6 A0A2Z2U8A4 A0A3Q3KIW6 A0A3Q3BT62 A0A0F8APH6 A0A1A7Y5C8 A0A147A0Y3 A0A1A8EWH3
S4PL92 A0A212EWQ2 H9JFK7 A0A0L7LP67 A0A087TFB1 E0VM17 A0A0K8UFB8 A0A034VRF0 A0A2H8TGM0 A0A0M3TCD7 A0A067QWC1 D2A1F0 U5ESE8 A0A2S2Q587 J9JPP1 A0A1Y1LC46 A0A2L2Y9W6 W8BSF8 A0A0T6AVT9 A0A131Y2G3 A0A147BUH3 T1P884 A0A182WNJ4 A0A1S3DA70 A0A1I8PVS7 A0A2M3YYS8 A0A2M3YYW6 A0A1B6GA14 V5H036 A0A182NY76 A0A2R5LBS4 Q7QK56 A0A3F2YSG6 A0A182XT68 A0A182VCH1 A0A182MDR8 Q8INB9-2 A0A0B4LIA3 B4QXQ5 B4HLD8 A0A293M499 B3P0Y2 H1ZY63 H1ZY64 Q8INB9 T1JL53 A0A0V1LT75 A0A0V0X785 A0A0V1PFC8 A0A1Y3EJB3 A0A0V0SNC6 A0A023GNP7 A0A1E1X356 A0A131YY67 A0A0V1C341 A0A224YYY4 L7MAG4 L7RA14 A0A1E1XRF4 A0A0V1N108 F5HR84 A0A0V1C2F7 A0A131XFF7 A0A0V1C2D6 A0A0V1FWK3 A0A0V1JFA2 A0A0V1AG58 A0A077YWW3 A0A0V0VUE6 A0A3Q0J4F5 A0A1D1VR66 A0A210Q3T4 A0A159ZKT0 A0A0V0UG04 A0A0V0UHG7 A0A0V1N1E2 A0A085NKI5 V9KNB7 A0A1S3JCH1 B5THM6 A0A0N5DJ40 A0A0S2GL00 W5K6L6 A0A2D0Q8N4 A0A3B4DV68 A0A0S7KZT5 E7EXT6 A0A2Z2U8A4 A0A3Q3KIW6 A0A3Q3BT62 A0A0F8APH6 A0A1A7Y5C8 A0A147A0Y3 A0A1A8EWH3
EC Number
2.7.11.1
Pubmed
20197069
26354079
23622113
22118469
19121390
26227816
+ More
20566863 25348373 26701614 24845553 18362917 19820115 28004739 26561354 24495485 29652888 25315136 12364791 14747013 17210077 8302573 7876156 10731132 12537572 12537569 9601646 10587646 10962553 11740943 11344272 11752451 12172554 11872800 14525946 12893776 15466161 15712201 15808505 15718470 18327897 22493059 24786828 24603715 25329475 29025897 12537568 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 28503490 26830274 28797301 25576852 23274741 29209593 22193391 28049606 27649274 28812685 25158315 24929829 24402279 26549179 25329095 23594743 25835551
20566863 25348373 26701614 24845553 18362917 19820115 28004739 26561354 24495485 29652888 25315136 12364791 14747013 17210077 8302573 7876156 10731132 12537572 12537569 9601646 10587646 10962553 11740943 11344272 11752451 12172554 11872800 14525946 12893776 15466161 15712201 15808505 15718470 18327897 22493059 24786828 24603715 25329475 29025897 12537568 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 28503490 26830274 28797301 25576852 23274741 29209593 22193391 28049606 27649274 28812685 25158315 24929829 24402279 26549179 25329095 23594743 25835551
EMBL
EU305740
ABY50539.1
NWSH01002510
PCG68106.1
JN399217
AEM63703.1
+ More
MG657021 AXZ96481.1 KQ458880 KPJ04674.1 KQ460154 KPJ17321.1 GAIX01004160 JAA88400.1 AGBW02011923 OWR45915.1 BABH01002405 BABH01002406 BABH01002407 JTDY01000430 KOB77215.1 KK114965 KFM63800.1 DS235286 EEB14423.1 GDHF01029189 GDHF01027048 GDHF01009039 GDHF01007074 GDHF01005983 JAI23125.1 JAI25266.1 JAI43275.1 JAI45240.1 JAI46331.1 KJ011125 GAKP01013076 GAKP01013075 GAKP01013074 GAKP01013073 AIC77786.1 JAC45876.1 GFXV01001386 MBW13191.1 KR075834 ALE20553.1 KK853289 KDR08900.1 KQ971338 EFA02105.1 GANO01003277 JAB56594.1 GGMS01003557 MBY72760.1 ABLF02033919 GEZM01060190 JAV71204.1 IAAA01011346 IAAA01011347 LAA04941.1 GAMC01010339 JAB96216.1 LJIG01022700 KRT79174.1 GEFM01002729 JAP73067.1 GEGO01000991 JAR94413.1 KA644799 AFP59428.1 GGFM01000665 MBW21416.1 GGFM01000711 MBW21462.1 GECZ01030783 GECZ01021084 GECZ01010492 GECZ01004955 JAS38986.1 JAS48685.1 JAS59277.1 JAS64814.1 GALX01000899 JAB67567.1 GGLE01002780 MBY06906.1 AAAB01008799 EAA03708.5 APCN01001995 APCN01001996 APCN01001997 AXCM01005381 Z26242 X83510 AE014297 AY069856 AHN57352.1 CM000364 EDX12729.1 CH480815 EDW41958.1 GFWV01010192 MAA34921.1 CH954181 EDV49101.1 FR870082 FR870084 FR870085 FR870090 FR870092 CCB63040.1 CCB63042.1 CCB63043.1 CCB63048.1 CCB63050.1 FR870081 FR870083 FR870086 FR870087 FR870088 FR870089 FR870091 FR870093 CCB63039.1 CCB63041.1 CCB63044.1 CCB63045.1 CCB63046.1 CCB63047.1 CCB63049.1 CCB63051.1 JH431868 JYDW01000006 KRZ62719.1 JYDK01000011 KRX83768.1 JYDM01000015 KRZ94928.1 LVZM01010870 OUC45065.1 JYDL01000001 KRX28218.1 GBBM01000838 JAC34580.1 GFAC01005505 JAT93683.1 GEDV01005065 JAP83492.1 JYDH01000001 KRY43484.1 GFPF01011670 MAA22816.1 GACK01004940 JAA60094.1 JX648548 AGC00787.1 GFAA01001539 JAU01896.1 JYDO01000018 KRZ77583.1 AB601888 BAK26531.1 KRY43482.1 GEFH01004205 JAP64376.1 KRY43481.1 JYDT01000023 KRY90257.1 JYDU01000020 JYDS01000008 KRX98585.1 KRZ33625.1 JYDQ01000003 KRY23530.1 HG805825 CDW52527.1 JYDN01000008 KRX66993.1 BDGG01000008 GAV02683.1 NEDP02005114 OWF43362.1 KX056492 AMY95372.1 JYDJ01000006 KRX50398.1 KRX50397.1 KRZ77582.1 KL363228 KL367491 KFD52402.1 KFD69981.1 JW867172 AFO99689.1 EU939752 ACH73234.1 KT362182 ALN96976.1 GBYX01191286 GBYX01191285 GBYX01191282 JAO82760.1 CU928197 KX966254 ATO58448.1 KQ041256 KKF28993.1 HADW01014163 HADX01003261 SBP25493.1 GCES01014100 JAR72223.1 HAEB01004958 SBQ51485.1
MG657021 AXZ96481.1 KQ458880 KPJ04674.1 KQ460154 KPJ17321.1 GAIX01004160 JAA88400.1 AGBW02011923 OWR45915.1 BABH01002405 BABH01002406 BABH01002407 JTDY01000430 KOB77215.1 KK114965 KFM63800.1 DS235286 EEB14423.1 GDHF01029189 GDHF01027048 GDHF01009039 GDHF01007074 GDHF01005983 JAI23125.1 JAI25266.1 JAI43275.1 JAI45240.1 JAI46331.1 KJ011125 GAKP01013076 GAKP01013075 GAKP01013074 GAKP01013073 AIC77786.1 JAC45876.1 GFXV01001386 MBW13191.1 KR075834 ALE20553.1 KK853289 KDR08900.1 KQ971338 EFA02105.1 GANO01003277 JAB56594.1 GGMS01003557 MBY72760.1 ABLF02033919 GEZM01060190 JAV71204.1 IAAA01011346 IAAA01011347 LAA04941.1 GAMC01010339 JAB96216.1 LJIG01022700 KRT79174.1 GEFM01002729 JAP73067.1 GEGO01000991 JAR94413.1 KA644799 AFP59428.1 GGFM01000665 MBW21416.1 GGFM01000711 MBW21462.1 GECZ01030783 GECZ01021084 GECZ01010492 GECZ01004955 JAS38986.1 JAS48685.1 JAS59277.1 JAS64814.1 GALX01000899 JAB67567.1 GGLE01002780 MBY06906.1 AAAB01008799 EAA03708.5 APCN01001995 APCN01001996 APCN01001997 AXCM01005381 Z26242 X83510 AE014297 AY069856 AHN57352.1 CM000364 EDX12729.1 CH480815 EDW41958.1 GFWV01010192 MAA34921.1 CH954181 EDV49101.1 FR870082 FR870084 FR870085 FR870090 FR870092 CCB63040.1 CCB63042.1 CCB63043.1 CCB63048.1 CCB63050.1 FR870081 FR870083 FR870086 FR870087 FR870088 FR870089 FR870091 FR870093 CCB63039.1 CCB63041.1 CCB63044.1 CCB63045.1 CCB63046.1 CCB63047.1 CCB63049.1 CCB63051.1 JH431868 JYDW01000006 KRZ62719.1 JYDK01000011 KRX83768.1 JYDM01000015 KRZ94928.1 LVZM01010870 OUC45065.1 JYDL01000001 KRX28218.1 GBBM01000838 JAC34580.1 GFAC01005505 JAT93683.1 GEDV01005065 JAP83492.1 JYDH01000001 KRY43484.1 GFPF01011670 MAA22816.1 GACK01004940 JAA60094.1 JX648548 AGC00787.1 GFAA01001539 JAU01896.1 JYDO01000018 KRZ77583.1 AB601888 BAK26531.1 KRY43482.1 GEFH01004205 JAP64376.1 KRY43481.1 JYDT01000023 KRY90257.1 JYDU01000020 JYDS01000008 KRX98585.1 KRZ33625.1 JYDQ01000003 KRY23530.1 HG805825 CDW52527.1 JYDN01000008 KRX66993.1 BDGG01000008 GAV02683.1 NEDP02005114 OWF43362.1 KX056492 AMY95372.1 JYDJ01000006 KRX50398.1 KRX50397.1 KRZ77582.1 KL363228 KL367491 KFD52402.1 KFD69981.1 JW867172 AFO99689.1 EU939752 ACH73234.1 KT362182 ALN96976.1 GBYX01191286 GBYX01191285 GBYX01191282 JAO82760.1 CU928197 KX966254 ATO58448.1 KQ041256 KKF28993.1 HADW01014163 HADX01003261 SBP25493.1 GCES01014100 JAR72223.1 HAEB01004958 SBQ51485.1
Proteomes
UP000218220
UP000053268
UP000053240
UP000007151
UP000005204
UP000037510
+ More
UP000054359 UP000009046 UP000027135 UP000007266 UP000007819 UP000095301 UP000075920 UP000079169 UP000095300 UP000075884 UP000007062 UP000075840 UP000076407 UP000075903 UP000075883 UP000000803 UP000000304 UP000001292 UP000008711 UP000054721 UP000054673 UP000054924 UP000243006 UP000054630 UP000054776 UP000054843 UP000054995 UP000054805 UP000054815 UP000054783 UP000030665 UP000054681 UP000186922 UP000242188 UP000055048 UP000085678 UP000046395 UP000018467 UP000221080 UP000261440 UP000000437 UP000261640 UP000264840 UP000265000
UP000054359 UP000009046 UP000027135 UP000007266 UP000007819 UP000095301 UP000075920 UP000079169 UP000095300 UP000075884 UP000007062 UP000075840 UP000076407 UP000075903 UP000075883 UP000000803 UP000000304 UP000001292 UP000008711 UP000054721 UP000054673 UP000054924 UP000243006 UP000054630 UP000054776 UP000054843 UP000054995 UP000054805 UP000054815 UP000054783 UP000030665 UP000054681 UP000186922 UP000242188 UP000055048 UP000085678 UP000046395 UP000018467 UP000221080 UP000261440 UP000000437 UP000261640 UP000264840 UP000265000
Interpro
IPR000961
AGC-kinase_C
+ More
IPR000719 Prot_kinase_dom
IPR017892 Pkinase_C
IPR011993 PH-like_dom_sf
IPR017441 Protein_kinase_ATP_BS
IPR039026 PH_PKB
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
IPR001849 PH_domain
IPR039029 RAC_gamma-like
IPR016024 ARM-type_fold
IPR013598 Exportin-1/Importin-b-like
IPR004299 MBOAT_fam
IPR000719 Prot_kinase_dom
IPR017892 Pkinase_C
IPR011993 PH-like_dom_sf
IPR017441 Protein_kinase_ATP_BS
IPR039026 PH_PKB
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
IPR001849 PH_domain
IPR039029 RAC_gamma-like
IPR016024 ARM-type_fold
IPR013598 Exportin-1/Importin-b-like
IPR004299 MBOAT_fam
Gene 3D
CDD
ProteinModelPortal
B0FC95
A0A2A4J8L6
G3LIS8
A0A385KNU3
A0A194QGX8
A0A194RMU6
+ More
S4PL92 A0A212EWQ2 H9JFK7 A0A0L7LP67 A0A087TFB1 E0VM17 A0A0K8UFB8 A0A034VRF0 A0A2H8TGM0 A0A0M3TCD7 A0A067QWC1 D2A1F0 U5ESE8 A0A2S2Q587 J9JPP1 A0A1Y1LC46 A0A2L2Y9W6 W8BSF8 A0A0T6AVT9 A0A131Y2G3 A0A147BUH3 T1P884 A0A182WNJ4 A0A1S3DA70 A0A1I8PVS7 A0A2M3YYS8 A0A2M3YYW6 A0A1B6GA14 V5H036 A0A182NY76 A0A2R5LBS4 Q7QK56 A0A3F2YSG6 A0A182XT68 A0A182VCH1 A0A182MDR8 Q8INB9-2 A0A0B4LIA3 B4QXQ5 B4HLD8 A0A293M499 B3P0Y2 H1ZY63 H1ZY64 Q8INB9 T1JL53 A0A0V1LT75 A0A0V0X785 A0A0V1PFC8 A0A1Y3EJB3 A0A0V0SNC6 A0A023GNP7 A0A1E1X356 A0A131YY67 A0A0V1C341 A0A224YYY4 L7MAG4 L7RA14 A0A1E1XRF4 A0A0V1N108 F5HR84 A0A0V1C2F7 A0A131XFF7 A0A0V1C2D6 A0A0V1FWK3 A0A0V1JFA2 A0A0V1AG58 A0A077YWW3 A0A0V0VUE6 A0A3Q0J4F5 A0A1D1VR66 A0A210Q3T4 A0A159ZKT0 A0A0V0UG04 A0A0V0UHG7 A0A0V1N1E2 A0A085NKI5 V9KNB7 A0A1S3JCH1 B5THM6 A0A0N5DJ40 A0A0S2GL00 W5K6L6 A0A2D0Q8N4 A0A3B4DV68 A0A0S7KZT5 E7EXT6 A0A2Z2U8A4 A0A3Q3KIW6 A0A3Q3BT62 A0A0F8APH6 A0A1A7Y5C8 A0A147A0Y3 A0A1A8EWH3
S4PL92 A0A212EWQ2 H9JFK7 A0A0L7LP67 A0A087TFB1 E0VM17 A0A0K8UFB8 A0A034VRF0 A0A2H8TGM0 A0A0M3TCD7 A0A067QWC1 D2A1F0 U5ESE8 A0A2S2Q587 J9JPP1 A0A1Y1LC46 A0A2L2Y9W6 W8BSF8 A0A0T6AVT9 A0A131Y2G3 A0A147BUH3 T1P884 A0A182WNJ4 A0A1S3DA70 A0A1I8PVS7 A0A2M3YYS8 A0A2M3YYW6 A0A1B6GA14 V5H036 A0A182NY76 A0A2R5LBS4 Q7QK56 A0A3F2YSG6 A0A182XT68 A0A182VCH1 A0A182MDR8 Q8INB9-2 A0A0B4LIA3 B4QXQ5 B4HLD8 A0A293M499 B3P0Y2 H1ZY63 H1ZY64 Q8INB9 T1JL53 A0A0V1LT75 A0A0V0X785 A0A0V1PFC8 A0A1Y3EJB3 A0A0V0SNC6 A0A023GNP7 A0A1E1X356 A0A131YY67 A0A0V1C341 A0A224YYY4 L7MAG4 L7RA14 A0A1E1XRF4 A0A0V1N108 F5HR84 A0A0V1C2F7 A0A131XFF7 A0A0V1C2D6 A0A0V1FWK3 A0A0V1JFA2 A0A0V1AG58 A0A077YWW3 A0A0V0VUE6 A0A3Q0J4F5 A0A1D1VR66 A0A210Q3T4 A0A159ZKT0 A0A0V0UG04 A0A0V0UHG7 A0A0V1N1E2 A0A085NKI5 V9KNB7 A0A1S3JCH1 B5THM6 A0A0N5DJ40 A0A0S2GL00 W5K6L6 A0A2D0Q8N4 A0A3B4DV68 A0A0S7KZT5 E7EXT6 A0A2Z2U8A4 A0A3Q3KIW6 A0A3Q3BT62 A0A0F8APH6 A0A1A7Y5C8 A0A147A0Y3 A0A1A8EWH3
PDB
3O96
E-value=1.14566e-110,
Score=1021
Ontologies
PATHWAY
04068
FoxO signaling pathway - Bombyx mori (domestic silkworm)
04140 Autophagy - animal - Bombyx mori (domestic silkworm)
04150 mTOR signaling pathway - Bombyx mori (domestic silkworm)
04213 Longevity regulating pathway - multiple species - Bombyx mori (domestic silkworm)
04933 AGE-RAGE signaling pathway in diabetic complications - Bombyx mori (domestic silkworm)
04140 Autophagy - animal - Bombyx mori (domestic silkworm)
04150 mTOR signaling pathway - Bombyx mori (domestic silkworm)
04213 Longevity regulating pathway - multiple species - Bombyx mori (domestic silkworm)
04933 AGE-RAGE signaling pathway in diabetic complications - Bombyx mori (domestic silkworm)
GO
GO:0004674
GO:0005524
GO:0043491
GO:0035556
GO:0006468
GO:0005737
GO:0005634
GO:0018105
GO:0032869
GO:0031104
GO:0007424
GO:0045793
GO:0007623
GO:0007525
GO:0008284
GO:1901215
GO:0043066
GO:1904263
GO:0043025
GO:0050773
GO:0005829
GO:0042632
GO:0035206
GO:0045886
GO:0005938
GO:0006629
GO:0090278
GO:0040014
GO:0048010
GO:0040018
GO:0007520
GO:0060292
GO:0010884
GO:0007427
GO:0008362
GO:0006915
GO:0005886
GO:0046622
GO:0035264
GO:0048477
GO:0030307
GO:0010897
GO:0048680
GO:0008286
GO:0006979
GO:0035091
GO:1904262
GO:0016021
GO:0004672
GO:0000155
GO:0000160
GO:0005216
GO:0006811
GO:0005622
GO:0006810
GO:0003824
PANTHER
Topology
Subcellular location
Cytoplasm
Cytosol
Cell membrane
Nucleus
Cytosol
Cell membrane
Nucleus
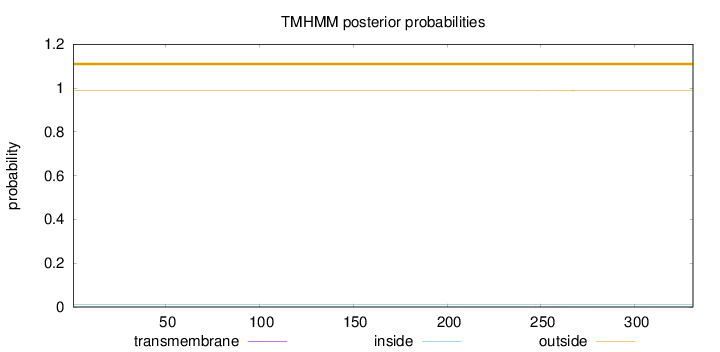
Length:
331
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01458
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01072
outside
1 - 331
Population Genetic Test Statistics
Pi
242.73269
Theta
197.764145
Tajima's D
0.735846
CLR
0.302281
CSRT
0.586870656467177
Interpretation
Uncertain
