Gene
KWMTBOMO11070 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008555
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_4-coumarate--CoA_ligase_1-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.056
Sequence
CDS
ATGTCGAATACATTGAGACGTACTGCACAACAATTGTTGAAGACACACAAGTATGCAACTGCAACATCAAGTATAAGGCAAAAAACAGTAGAAAAAAACATTCTGACATCTGTCTACAAGGATATTCCTGTGACAAATATGACTATCAATGATTTTGTTTGGCAAAATTTGGATAGATGGCCCGACAAGACTGCTACAGTATGCGCGCTGACCGGTCACGGGTACACTTACGCCCAAACGCACAGGATGTCGGTAACATTCGCAGCGTCCTTACGATCGAAACTGAAACTGAAGAATGACGACAAAGTGGCGGTTATCTTGCCGAATTTACCAGAATATCCCTGTACATTGCTGGGCATATTGCAGGCTGGCTGTGTTGCCAGCATGATGAACCCAGTTTACACGTCTCATGAATTAAAACGACAATTGGAGTTGATCGATTGTAAAGCTATCGTCAGTTCCAAAATATCGTACGCGAACATAAAACAGGCACTGGACGAGTTGAAAGTCAACGTGCCTATAATACTAGTCGACAACGAGGGTCTGCCCGAGGGCGCGATAAAATTCGCCGAACTAGCCGAAGACTTCAACGTCGACATTGACTGTTTGAAGTCCGTTCAGAGGGGTCCCAAAGACGTCGCGATCCTGCCGTTCTCCAGTGGAACCACGGGCTTCCCCAAGGCCGTGGTTCTCGATCACGGAGGCGTTGTCGCCTTGAACCAAATGATAATAGACCCGGAGATTGTTGTGATAGAAGAGACCACCGCAACACACCAGTCAGTGTTGCCAGCGATCTTGCCGTTTTTCCACATATTCGGTTTCAATTCCCTAATGATGAACCAAATGGCGCAGGGCGTGAAATTAGTCACGATGCCGTATTTCAAACCGGATCTTTTCCTGCAGACCATCGTGAAGTACAAGGCGGAAGTGCTCTTCATTGTTCCGCCGATGGTGGTATTCCTGGGTAAGCATCCGGCGGTTAAACCTGAACATCTGGCCGCCGTCAAGGGCATGATCAGCGGAGCCGCTCCGCTCTCGGCCGCGGATGCTGCGGCTGTATTGGAAAAGAACAAAAACATAATATTCCGGCAAGGTTACGGCCTGACCGAGACCAATGGAGCCGTCACTGTCGGTAGGAATGATGACACGAACCACGCGGCCGTTGGACACGTGCTAGGCAGCTGCGAGGTCAAGGTAGCTGACGTCAACACGCAGGAAGCATTGGGCGCGGGGCAGGAAGGCGAGATCTGGTGTCGCGGCCCAAACCTGATGTCCGGTTATTATCTAGACGAAAAGTCCACCAGGGAGGTCATGACGGAGGATGGCTGGTACAAAACCGGAGACATTGGGAAGTACGACGAGAACAAATATCTGTACGTCACGGATAGACTGAAGGAACTCATCAAGGTCAAAGGTTTCCAAGTACCGCCTGCTGAATTGGAGACCGTCATCAGAACGCACTCCAAGGTTCTAGACTGCGCGGTCCTGGGCATACCGGACCCCGTAACAGGGGAAGTACCGAAGGCCTTTATAGTCCCTCAACAGGGTCAGACCCTCGACGCTGATGAGTTGTTAGAGTACGTCAACAGTAAAGTGGCCGTCTTCAAGAAGATCAAGGAAGTCCAGTTCGTGGAAGAAATACCCAAGAATTCGGCAGAGTTATATGTATAA
Protein
MSNTLRRTAQQLLKTHKYATATSSIRQKTVEKNILTSVYKDIPVTNMTINDFVWQNLDRWPDKTATVCALTGHGYTYAQTHRMSVTFAASLRSKLKLKNDDKVAVILPNLPEYPCTLLGILQAGCVASMMNPVYTSHELKRQLELIDCKAIVSSKISYANIKQALDELKVNVPIILVDNEGLPEGAIKFAELAEDFNVDIDCLKSVQRGPKDVAILPFSSGTTGFPKAVVLDHGGVVALNQMIIDPEIVVIEETTATHQSVLPAILPFFHIFGFNSLMMNQMAQGVKLVTMPYFKPDLFLQTIVKYKAEVLFIVPPMVVFLGKHPAVKPEHLAAVKGMISGAAPLSAADAAAVLEKNKNIIFRQGYGLTETNGAVTVGRNDDTNHAAVGHVLGSCEVKVADVNTQEALGAGQEGEIWCRGPNLMSGYYLDEKSTREVMTEDGWYKTGDIGKYDENKYLYVTDRLKELIKVKGFQVPPAELETVIRTHSKVLDCAVLGIPDPVTGEVPKAFIVPQQGQTLDADELLEYVNSKVAVFKKIKEVQFVEEIPKNSAELYV
Summary
Uniprot
H9JGA8
A0A2A4K2K9
A0A2H1VTI1
A0A212ELC6
A0A194RII1
A0A2A4K2X8
+ More
A0A194QI42 A0A2H1V6M1 A0A212ELD3 H9JGA9 A0A2A4K3A4 A0A1Q3FKE9 A0A1Q3FJV1 A0A1Q3FJY7 A0A3S2NX79 B0W6R4 A0A182NR50 A0A182JN78 A0A2H1WC02 A0A182K9W2 A0A182UUK9 A0A182TPK3 Q7QCM8 A0A182R455 A0A182LE43 A0A182HS75 A0A182XAD5 A0A182Y402 A0A1L8DYU5 A0A182PUZ1 A0A1L8DYX6 A0A182VZU2 A0A182LUB0 A0A1L8DYT3 A0A182FQZ2 A0A2M4A7J6 Q17P68 A0A2M4A9F3 A0A182QDT5 W5JKB2 A0A2M4BIF8 A0A2M3ZGD1 A0A2M4BHT0 A0A2H1V6K9 A0A2M4BKZ5 A0A1B0GLA5 A0A3S2LHM1 A0A2J7QAA1 T1PER9 A0A1I8N0U5 A0A182H784 U5EJG4 A0A194RIT5 A0A2A4IY84 A0A0A1WUA9 B3MXL5 A0A194QGE2 A0A0L0CJM9 B4L257 Q29IR9 B4MTA9 H9JGB0 B4JL04 A0A2J7RB04 A0A2J7RB11 A0A3B0JZ59 A0A0N1IT33 B4M8E6 A0A0C9R6E2 A0A2J7QWJ4 A0A1I8QB84 Q9VXZ8 B4PW91 A0A0M5J603 A0A067R080 A0A1W4VFX1 A0A034VMV0 D2A1H8 B3NUW6 A0A2A4K1W8 D2A1H7 A0A194QGQ5 A0A195D8L9 A0A069DUU3 W8AQQ0 A0A195D1W7 A0A026WGV0 A0A195D8N5 A0A0K8SSC0 A0A146L4D3 B4R4X2 A0A0L7R6L2 A0A151WYA4 A0A0N7Z8U2 T1H7X8 A0A2H1VTM1 A0A158ND07 V5GTJ7 E2A045
A0A194QI42 A0A2H1V6M1 A0A212ELD3 H9JGA9 A0A2A4K3A4 A0A1Q3FKE9 A0A1Q3FJV1 A0A1Q3FJY7 A0A3S2NX79 B0W6R4 A0A182NR50 A0A182JN78 A0A2H1WC02 A0A182K9W2 A0A182UUK9 A0A182TPK3 Q7QCM8 A0A182R455 A0A182LE43 A0A182HS75 A0A182XAD5 A0A182Y402 A0A1L8DYU5 A0A182PUZ1 A0A1L8DYX6 A0A182VZU2 A0A182LUB0 A0A1L8DYT3 A0A182FQZ2 A0A2M4A7J6 Q17P68 A0A2M4A9F3 A0A182QDT5 W5JKB2 A0A2M4BIF8 A0A2M3ZGD1 A0A2M4BHT0 A0A2H1V6K9 A0A2M4BKZ5 A0A1B0GLA5 A0A3S2LHM1 A0A2J7QAA1 T1PER9 A0A1I8N0U5 A0A182H784 U5EJG4 A0A194RIT5 A0A2A4IY84 A0A0A1WUA9 B3MXL5 A0A194QGE2 A0A0L0CJM9 B4L257 Q29IR9 B4MTA9 H9JGB0 B4JL04 A0A2J7RB04 A0A2J7RB11 A0A3B0JZ59 A0A0N1IT33 B4M8E6 A0A0C9R6E2 A0A2J7QWJ4 A0A1I8QB84 Q9VXZ8 B4PW91 A0A0M5J603 A0A067R080 A0A1W4VFX1 A0A034VMV0 D2A1H8 B3NUW6 A0A2A4K1W8 D2A1H7 A0A194QGQ5 A0A195D8L9 A0A069DUU3 W8AQQ0 A0A195D1W7 A0A026WGV0 A0A195D8N5 A0A0K8SSC0 A0A146L4D3 B4R4X2 A0A0L7R6L2 A0A151WYA4 A0A0N7Z8U2 T1H7X8 A0A2H1VTM1 A0A158ND07 V5GTJ7 E2A045
Pubmed
19121390
22118469
26354079
12364791
14747013
17210077
+ More
20966253 25244985 17510324 20920257 23761445 25315136 26483478 25830018 17994087 26108605 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 24845553 25348373 18362917 19820115 26334808 24495485 24508170 30249741 26823975 27129103 21347285 20798317
20966253 25244985 17510324 20920257 23761445 25315136 26483478 25830018 17994087 26108605 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 24845553 25348373 18362917 19820115 26334808 24495485 24508170 30249741 26823975 27129103 21347285 20798317
EMBL
BABH01002391
BABH01002392
NWSH01000221
PCG78256.1
ODYU01004348
SOQ44127.1
+ More
AGBW02014086 OWR42279.1 KQ460154 KPJ17249.1 PCG78254.1 KQ458880 KPJ04600.1 ODYU01000947 SOQ36467.1 OWR42277.1 BABH01002393 BABH01002394 PCG78253.1 GFDL01007082 JAV27963.1 GFDL01007188 JAV27857.1 GFDL01007104 JAV27941.1 RSAL01000117 RVE46830.1 DS231849 EDS36895.1 ODYU01007521 SOQ50362.1 AAAB01008859 EAA08143.4 APCN01000246 GFDF01002456 JAV11628.1 GFDF01002455 JAV11629.1 AXCM01000143 GFDF01002454 JAV11630.1 GGFK01003456 MBW36777.1 CH477193 EAT48565.1 GGFK01004041 MBW37362.1 AXCN02001476 ADMH02001265 ETN63325.1 GGFJ01003457 MBW52598.1 GGFM01006831 MBW27582.1 GGFJ01003456 MBW52597.1 SOQ36468.1 GGFJ01004327 MBW53468.1 AJWK01034692 AJWK01034693 RVE46823.1 NEVH01016331 PNF25515.1 KA647194 AFP61823.1 JXUM01027982 JXUM01027983 JXUM01027984 KQ560806 KXJ80822.1 GANO01002244 JAB57627.1 KPJ17250.1 NWSH01005551 PCG64130.1 GBXI01011870 GBXI01007925 JAD02422.1 JAD06367.1 CH902630 EDV38480.2 KPJ04603.1 JRES01000394 KNC31694.1 CH933810 EDW06797.2 CH379063 EAL32584.2 CH963851 EDW75348.2 BABH01002396 CH916370 EDW00257.1 NEVH01006564 PNF38010.1 PNF38011.1 OUUW01000003 SPP78666.1 KQ435896 KOX69252.1 CH940653 EDW62422.2 GBYB01002856 GBYB01011889 JAG72623.1 JAG81656.1 NEVH01009765 PNF32957.1 AF145610 AE014298 BT120049 AAD38585.1 AAF48408.2 ADA53588.1 ADV37672.1 AFH07387.1 AHN59727.1 CM000162 EDX01722.1 CP012528 ALC48836.1 KK852804 KDR16255.1 GAKP01015174 GAKP01015173 JAC43779.1 KQ971338 EFA01537.1 CH954180 EDV47064.1 PCG78255.1 EFA01538.1 KPJ04599.1 KQ981135 KYN09240.1 GBGD01001041 JAC87848.1 GAMC01015390 JAB91165.1 KQ977012 KYN06394.1 KK107213 QOIP01000007 EZA55292.1 RLU20572.1 KYN09238.1 GBRD01010094 JAG55730.1 GDHC01018826 GDHC01015238 JAP99802.1 JAQ03391.1 CM000366 EDX17929.1 KQ414646 KOC66391.1 KQ982650 KYQ52868.1 GDKW01002395 JAI54200.1 ACPB03018676 SOQ44128.1 ADTU01012156 GALX01004903 JAB63563.1 GL435551 EFN73178.1
AGBW02014086 OWR42279.1 KQ460154 KPJ17249.1 PCG78254.1 KQ458880 KPJ04600.1 ODYU01000947 SOQ36467.1 OWR42277.1 BABH01002393 BABH01002394 PCG78253.1 GFDL01007082 JAV27963.1 GFDL01007188 JAV27857.1 GFDL01007104 JAV27941.1 RSAL01000117 RVE46830.1 DS231849 EDS36895.1 ODYU01007521 SOQ50362.1 AAAB01008859 EAA08143.4 APCN01000246 GFDF01002456 JAV11628.1 GFDF01002455 JAV11629.1 AXCM01000143 GFDF01002454 JAV11630.1 GGFK01003456 MBW36777.1 CH477193 EAT48565.1 GGFK01004041 MBW37362.1 AXCN02001476 ADMH02001265 ETN63325.1 GGFJ01003457 MBW52598.1 GGFM01006831 MBW27582.1 GGFJ01003456 MBW52597.1 SOQ36468.1 GGFJ01004327 MBW53468.1 AJWK01034692 AJWK01034693 RVE46823.1 NEVH01016331 PNF25515.1 KA647194 AFP61823.1 JXUM01027982 JXUM01027983 JXUM01027984 KQ560806 KXJ80822.1 GANO01002244 JAB57627.1 KPJ17250.1 NWSH01005551 PCG64130.1 GBXI01011870 GBXI01007925 JAD02422.1 JAD06367.1 CH902630 EDV38480.2 KPJ04603.1 JRES01000394 KNC31694.1 CH933810 EDW06797.2 CH379063 EAL32584.2 CH963851 EDW75348.2 BABH01002396 CH916370 EDW00257.1 NEVH01006564 PNF38010.1 PNF38011.1 OUUW01000003 SPP78666.1 KQ435896 KOX69252.1 CH940653 EDW62422.2 GBYB01002856 GBYB01011889 JAG72623.1 JAG81656.1 NEVH01009765 PNF32957.1 AF145610 AE014298 BT120049 AAD38585.1 AAF48408.2 ADA53588.1 ADV37672.1 AFH07387.1 AHN59727.1 CM000162 EDX01722.1 CP012528 ALC48836.1 KK852804 KDR16255.1 GAKP01015174 GAKP01015173 JAC43779.1 KQ971338 EFA01537.1 CH954180 EDV47064.1 PCG78255.1 EFA01538.1 KPJ04599.1 KQ981135 KYN09240.1 GBGD01001041 JAC87848.1 GAMC01015390 JAB91165.1 KQ977012 KYN06394.1 KK107213 QOIP01000007 EZA55292.1 RLU20572.1 KYN09238.1 GBRD01010094 JAG55730.1 GDHC01018826 GDHC01015238 JAP99802.1 JAQ03391.1 CM000366 EDX17929.1 KQ414646 KOC66391.1 KQ982650 KYQ52868.1 GDKW01002395 JAI54200.1 ACPB03018676 SOQ44128.1 ADTU01012156 GALX01004903 JAB63563.1 GL435551 EFN73178.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000283053
+ More
UP000002320 UP000075884 UP000075880 UP000075881 UP000075903 UP000075902 UP000007062 UP000075900 UP000075882 UP000075840 UP000076407 UP000076408 UP000075885 UP000075920 UP000075883 UP000069272 UP000008820 UP000075886 UP000000673 UP000092461 UP000235965 UP000095301 UP000069940 UP000249989 UP000007801 UP000037069 UP000009192 UP000001819 UP000007798 UP000001070 UP000268350 UP000053105 UP000008792 UP000095300 UP000000803 UP000002282 UP000092553 UP000027135 UP000192221 UP000007266 UP000008711 UP000078492 UP000078542 UP000053097 UP000279307 UP000000304 UP000053825 UP000075809 UP000015103 UP000005205 UP000000311
UP000002320 UP000075884 UP000075880 UP000075881 UP000075903 UP000075902 UP000007062 UP000075900 UP000075882 UP000075840 UP000076407 UP000076408 UP000075885 UP000075920 UP000075883 UP000069272 UP000008820 UP000075886 UP000000673 UP000092461 UP000235965 UP000095301 UP000069940 UP000249989 UP000007801 UP000037069 UP000009192 UP000001819 UP000007798 UP000001070 UP000268350 UP000053105 UP000008792 UP000095300 UP000000803 UP000002282 UP000092553 UP000027135 UP000192221 UP000007266 UP000008711 UP000078492 UP000078542 UP000053097 UP000279307 UP000000304 UP000053825 UP000075809 UP000015103 UP000005205 UP000000311
Interpro
Gene 3D
ProteinModelPortal
H9JGA8
A0A2A4K2K9
A0A2H1VTI1
A0A212ELC6
A0A194RII1
A0A2A4K2X8
+ More
A0A194QI42 A0A2H1V6M1 A0A212ELD3 H9JGA9 A0A2A4K3A4 A0A1Q3FKE9 A0A1Q3FJV1 A0A1Q3FJY7 A0A3S2NX79 B0W6R4 A0A182NR50 A0A182JN78 A0A2H1WC02 A0A182K9W2 A0A182UUK9 A0A182TPK3 Q7QCM8 A0A182R455 A0A182LE43 A0A182HS75 A0A182XAD5 A0A182Y402 A0A1L8DYU5 A0A182PUZ1 A0A1L8DYX6 A0A182VZU2 A0A182LUB0 A0A1L8DYT3 A0A182FQZ2 A0A2M4A7J6 Q17P68 A0A2M4A9F3 A0A182QDT5 W5JKB2 A0A2M4BIF8 A0A2M3ZGD1 A0A2M4BHT0 A0A2H1V6K9 A0A2M4BKZ5 A0A1B0GLA5 A0A3S2LHM1 A0A2J7QAA1 T1PER9 A0A1I8N0U5 A0A182H784 U5EJG4 A0A194RIT5 A0A2A4IY84 A0A0A1WUA9 B3MXL5 A0A194QGE2 A0A0L0CJM9 B4L257 Q29IR9 B4MTA9 H9JGB0 B4JL04 A0A2J7RB04 A0A2J7RB11 A0A3B0JZ59 A0A0N1IT33 B4M8E6 A0A0C9R6E2 A0A2J7QWJ4 A0A1I8QB84 Q9VXZ8 B4PW91 A0A0M5J603 A0A067R080 A0A1W4VFX1 A0A034VMV0 D2A1H8 B3NUW6 A0A2A4K1W8 D2A1H7 A0A194QGQ5 A0A195D8L9 A0A069DUU3 W8AQQ0 A0A195D1W7 A0A026WGV0 A0A195D8N5 A0A0K8SSC0 A0A146L4D3 B4R4X2 A0A0L7R6L2 A0A151WYA4 A0A0N7Z8U2 T1H7X8 A0A2H1VTM1 A0A158ND07 V5GTJ7 E2A045
A0A194QI42 A0A2H1V6M1 A0A212ELD3 H9JGA9 A0A2A4K3A4 A0A1Q3FKE9 A0A1Q3FJV1 A0A1Q3FJY7 A0A3S2NX79 B0W6R4 A0A182NR50 A0A182JN78 A0A2H1WC02 A0A182K9W2 A0A182UUK9 A0A182TPK3 Q7QCM8 A0A182R455 A0A182LE43 A0A182HS75 A0A182XAD5 A0A182Y402 A0A1L8DYU5 A0A182PUZ1 A0A1L8DYX6 A0A182VZU2 A0A182LUB0 A0A1L8DYT3 A0A182FQZ2 A0A2M4A7J6 Q17P68 A0A2M4A9F3 A0A182QDT5 W5JKB2 A0A2M4BIF8 A0A2M3ZGD1 A0A2M4BHT0 A0A2H1V6K9 A0A2M4BKZ5 A0A1B0GLA5 A0A3S2LHM1 A0A2J7QAA1 T1PER9 A0A1I8N0U5 A0A182H784 U5EJG4 A0A194RIT5 A0A2A4IY84 A0A0A1WUA9 B3MXL5 A0A194QGE2 A0A0L0CJM9 B4L257 Q29IR9 B4MTA9 H9JGB0 B4JL04 A0A2J7RB04 A0A2J7RB11 A0A3B0JZ59 A0A0N1IT33 B4M8E6 A0A0C9R6E2 A0A2J7QWJ4 A0A1I8QB84 Q9VXZ8 B4PW91 A0A0M5J603 A0A067R080 A0A1W4VFX1 A0A034VMV0 D2A1H8 B3NUW6 A0A2A4K1W8 D2A1H7 A0A194QGQ5 A0A195D8L9 A0A069DUU3 W8AQQ0 A0A195D1W7 A0A026WGV0 A0A195D8N5 A0A0K8SSC0 A0A146L4D3 B4R4X2 A0A0L7R6L2 A0A151WYA4 A0A0N7Z8U2 T1H7X8 A0A2H1VTM1 A0A158ND07 V5GTJ7 E2A045
PDB
3TSY
E-value=1.06781e-76,
Score=731
Ontologies
GO
Topology
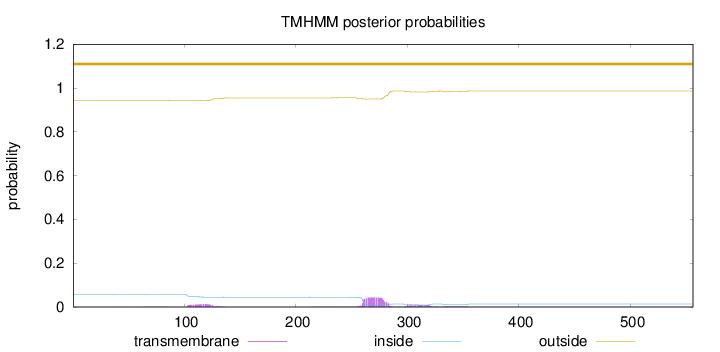
Length:
556
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.47408
Exp number, first 60 AAs:
0.00165
Total prob of N-in:
0.05796
outside
1 - 556
Population Genetic Test Statistics
Pi
173.044639
Theta
162.622635
Tajima's D
0.326752
CLR
0.240548
CSRT
0.469426528673566
Interpretation
Uncertain
