Pre Gene Modal
BGIBMGA008554
Annotation
PREDICTED:_mitochondrial_genome_maintenance_exonuclease_1-like_[Amyelois_transitella]
Full name
Mitochondrial genome maintenance exonuclease 1
Location in the cell
Mitochondrial Reliability : 1.943
Sequence
CDS
ATGTGGCAAAATACAAGGCACTGTTTGAATATTTTTGGAGTTCTTTTAAATAACACAGCTATACGTGGCTTAAAAACATCTGTAGTATGTGGTACAGCCAAAAGTTCACCAGCGCAACCTGGATATCCCAGTGAAGATAAAATCTTGCAGATAAAACACCACAGAAATGATTATGCTGAACAATACCCATCGGTGACACTAATATTAAATAAAACAATGACTGAAGAGTCTAGGTTGGCTTTGGAGAAGTGGAAACAAGAGAGGATTGCCGAAATGGGAGTTGAAGAATTTAATAGATTTTACCAAGCTCAAATGGCAGTCGGTACCAAGTTTCACAACACTTTAAAGGATTACTTCACTCAACCTCGTAGCCAGCTTCGTATAGAGAAGGAAGTGGAGGGTGTATGGGTGTCTGTAGCAGATGTGTTGAAAAGTATATCTTCTCCAAAAGCCATAGAATCGAATGTTGTACACCCCGTACTGAAGTATAGGGGAATTTTTGATGCCATTGCTGATTATGAAGACAAACCGACTTTAATCGAGTGGAAGAAGTCTGATAAGCCGAGGAAGTCGATAGCCCTGACCTACGACAACCCGGTACAGCTGGCCGCTTACTTCGGTGCGGTCTGCAACGACCTCAACTACAAGCACATGAAGGTCCGCGACGCGCTGCTCGTCATAGCGTACACGGACGGCTCGAAGGCGGACGTCTTCCACCTGTCCACCGACAAGCTGAGGGAGCACTGGGCGCAGTGGCTGATCCGACTCGAGGAATACATAAACAAGCACGGCAAAGATTCCGAGAAGTTCCTGAAAGGCGGTAAAAGGTTGTTTGAGGTGGAAATCGGAAATCTCCAGTAA
Protein
MWQNTRHCLNIFGVLLNNTAIRGLKTSVVCGTAKSSPAQPGYPSEDKILQIKHHRNDYAEQYPSVTLILNKTMTEESRLALEKWKQERIAEMGVEEFNRFYQAQMAVGTKFHNTLKDYFTQPRSQLRIEKEVEGVWVSVADVLKSISSPKAIESNVVHPVLKYRGIFDAIADYEDKPTLIEWKKSDKPRKSIALTYDNPVQLAAYFGAVCNDLNYKHMKVRDALLVIAYTDGSKADVFHLSTDKLREHWAQWLIRLEEYINKHGKDSEKFLKGGKRLFEVEIGNLQ
Summary
Description
Metal-dependent single-stranded DNA (ssDNA) exonuclease involved in mitochondrial genome maintenance.
Metal-dependent single-stranded DNA (ssDNA) exonuclease involved in mitochondrial genome maintenance. Has preference for 5'-3' exonuclease activity. Necessary for maintenance of proper 7S DNA levels. Probably involved in mitochondrial DNA (mtDNA) repair.
Metal-dependent single-stranded DNA (ssDNA) exonuclease involved in mitochondrial genome maintenance. Has preference for 5'-3' exonuclease activity but is also capable of endoduclease activity on linear substrates. Necessary for maintenance of proper 7S DNA levels. Probably involved in mitochondrial DNA (mtDNA) repair, possibly via the processing of displaced DNA containing Okazaki fragments during RNA-primed DNA synthesis on the lagging strand or via processing of DNA flaps during long-patch base excision repair.
Metal-dependent single-stranded DNA (ssDNA) exonuclease involved in mitochondrial genome maintenance. Has preference for 5'-3' exonuclease activity. Necessary for maintenance of proper 7S DNA levels. Probably involved in mitochondrial DNA (mtDNA) repair.
Metal-dependent single-stranded DNA (ssDNA) exonuclease involved in mitochondrial genome maintenance. Has preference for 5'-3' exonuclease activity but is also capable of endoduclease activity on linear substrates. Necessary for maintenance of proper 7S DNA levels. Probably involved in mitochondrial DNA (mtDNA) repair, possibly via the processing of displaced DNA containing Okazaki fragments during RNA-primed DNA synthesis on the lagging strand or via processing of DNA flaps during long-patch base excision repair.
Similarity
Belongs to the MGME1 family.
Feature
chain Mitochondrial genome maintenance exonuclease 1
Uniprot
H9JGA7
A0A212ELA2
A0A194QM96
A0A1E1WAI0
A0A1E1WG20
A0A194RIH7
+ More
A0A3S2LY56 A0A0L7LAH9 A0A067RLA5 A0A0V0GCJ9 A0A2P8YGF1 A0A3B1JXT4 A0A224YNK8 A0A023G7P1 T1JCQ7 A0A2I4B122 A0A131XJL8 A0A1A7X9A2 A0A2U3YX08 A0A3P8UAN4 A0A0C9SC69 A0A3Q3CCN1 A0A3P8R923 A0A3P9BQB3 A0A3Q1EY05 A0A3B4GTB3 A0A3P9NJL6 A0A3Q1EA08 B7Q3F6 A0A315VR18 A0A131YR72 A0A3Q3IW50 I3JUQ2 A0A3Q3G760 A0A3M0J414 A0A3Q0RI01 A0A0S7HKA3 G1LF86 A0A3B4CKU8 A0A3L8SR64 D2HRE0 A0A131XVZ4 A0A3Q7PFV5 M3WRB4 A0A3B3YJN8 A0A2U3W7L1 H0Z676 A0A3Q7TSF4 G3NLE2 G3NLE4 V5HWA4 Q2WBW2 A0A3N0YVD1 A0A3Q3VVV1 E0VLE6 A0A087YGY6 A0A3B3TUG2 A0A3B5LRB9 A0A218V7F6 A0A3Q1AR02 A0A1A8JSV9 F4X1J1 G3GVW8 A0A1A8F434 W5Q9V5 K9IRU0 A0A384DML2 A0A3Q7WX20 A0A3Q3NA22 A0A2U9CCB9 U3JYR3 L8I3R8 B8A4D5 A0A3Q2HH00 Q1LZC4 A0A3B5R240 A0A2Y9IVI3 A0A1W4Z4I0 A0A1A8BCG0 G9L2D3 F6Q6G1 G3X2I1 A0A3B5A2M4 A0A1A8V9R6 A0A1A6HEX0
A0A3S2LY56 A0A0L7LAH9 A0A067RLA5 A0A0V0GCJ9 A0A2P8YGF1 A0A3B1JXT4 A0A224YNK8 A0A023G7P1 T1JCQ7 A0A2I4B122 A0A131XJL8 A0A1A7X9A2 A0A2U3YX08 A0A3P8UAN4 A0A0C9SC69 A0A3Q3CCN1 A0A3P8R923 A0A3P9BQB3 A0A3Q1EY05 A0A3B4GTB3 A0A3P9NJL6 A0A3Q1EA08 B7Q3F6 A0A315VR18 A0A131YR72 A0A3Q3IW50 I3JUQ2 A0A3Q3G760 A0A3M0J414 A0A3Q0RI01 A0A0S7HKA3 G1LF86 A0A3B4CKU8 A0A3L8SR64 D2HRE0 A0A131XVZ4 A0A3Q7PFV5 M3WRB4 A0A3B3YJN8 A0A2U3W7L1 H0Z676 A0A3Q7TSF4 G3NLE2 G3NLE4 V5HWA4 Q2WBW2 A0A3N0YVD1 A0A3Q3VVV1 E0VLE6 A0A087YGY6 A0A3B3TUG2 A0A3B5LRB9 A0A218V7F6 A0A3Q1AR02 A0A1A8JSV9 F4X1J1 G3GVW8 A0A1A8F434 W5Q9V5 K9IRU0 A0A384DML2 A0A3Q7WX20 A0A3Q3NA22 A0A2U9CCB9 U3JYR3 L8I3R8 B8A4D5 A0A3Q2HH00 Q1LZC4 A0A3B5R240 A0A2Y9IVI3 A0A1W4Z4I0 A0A1A8BCG0 G9L2D3 F6Q6G1 G3X2I1 A0A3B5A2M4 A0A1A8V9R6 A0A1A6HEX0
EC Number
3.1.-.-
Pubmed
EMBL
BABH01002387
AGBW02014086
OWR42280.1
KQ458880
KPJ04596.1
GDQN01007127
+ More
GDQN01003913 JAT83927.1 JAT87141.1 GDQN01005198 GDQN01004787 GDQN01004453 JAT85856.1 JAT86267.1 JAT86601.1 KQ460154 KPJ17244.1 RSAL01000117 RVE46834.1 JTDY01001979 KOB72415.1 KK852559 KDR21395.1 GECL01000283 JAP05841.1 PYGN01000614 PSN43329.1 GFPF01004418 MAA15564.1 GBBM01005207 JAC30211.1 JH432074 GEFH01000827 JAP67754.1 HADW01013118 HADX01008222 SBP14518.1 GBZX01001508 JAG91232.1 ABJB010921702 DS849458 EEC13378.1 NHOQ01001229 PWA25621.1 GEDV01007512 JAP81045.1 AERX01006458 QRBI01000208 RMB93643.1 GBYX01439831 JAO41527.1 ACTA01153874 ACTA01161873 QUSF01000008 RLW07020.1 GL193216 EFB28263.1 GEFM01006035 JAP69761.1 AANG04003379 ABQF01122982 GANP01005755 JAB78713.1 AM114790 CAJ38815.1 RJVU01026577 ROL49668.1 DS235271 EEB14202.1 AYCK01002128 MUZQ01000035 OWK61849.1 HAED01016181 HAEE01002957 SBR22977.1 GL888529 EGI59719.1 JH000044 EGV95405.1 HAEB01006994 HAEC01016191 SBQ53521.1 AMGL01024928 GABZ01002102 JAA51423.1 CP026257 AWP14215.1 AGTO01022007 JH882323 ELR49917.1 BX005372 BC116086 AAI16087.1 HADZ01000662 SBP64603.1 AEYP01035840 AEYP01035841 JP022714 AES11312.1 AEFK01029543 AEFK01029544 HAEJ01016648 SBS57105.1 LZPO01034865 OBS76796.1
GDQN01003913 JAT83927.1 JAT87141.1 GDQN01005198 GDQN01004787 GDQN01004453 JAT85856.1 JAT86267.1 JAT86601.1 KQ460154 KPJ17244.1 RSAL01000117 RVE46834.1 JTDY01001979 KOB72415.1 KK852559 KDR21395.1 GECL01000283 JAP05841.1 PYGN01000614 PSN43329.1 GFPF01004418 MAA15564.1 GBBM01005207 JAC30211.1 JH432074 GEFH01000827 JAP67754.1 HADW01013118 HADX01008222 SBP14518.1 GBZX01001508 JAG91232.1 ABJB010921702 DS849458 EEC13378.1 NHOQ01001229 PWA25621.1 GEDV01007512 JAP81045.1 AERX01006458 QRBI01000208 RMB93643.1 GBYX01439831 JAO41527.1 ACTA01153874 ACTA01161873 QUSF01000008 RLW07020.1 GL193216 EFB28263.1 GEFM01006035 JAP69761.1 AANG04003379 ABQF01122982 GANP01005755 JAB78713.1 AM114790 CAJ38815.1 RJVU01026577 ROL49668.1 DS235271 EEB14202.1 AYCK01002128 MUZQ01000035 OWK61849.1 HAED01016181 HAEE01002957 SBR22977.1 GL888529 EGI59719.1 JH000044 EGV95405.1 HAEB01006994 HAEC01016191 SBQ53521.1 AMGL01024928 GABZ01002102 JAA51423.1 CP026257 AWP14215.1 AGTO01022007 JH882323 ELR49917.1 BX005372 BC116086 AAI16087.1 HADZ01000662 SBP64603.1 AEYP01035840 AEYP01035841 JP022714 AES11312.1 AEFK01029543 AEFK01029544 HAEJ01016648 SBS57105.1 LZPO01034865 OBS76796.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000053240
UP000283053
UP000037510
+ More
UP000027135 UP000245037 UP000018467 UP000192220 UP000245341 UP000265080 UP000264840 UP000265100 UP000265160 UP000257200 UP000261460 UP000242638 UP000001555 UP000261600 UP000005207 UP000264800 UP000269221 UP000261340 UP000008912 UP000261440 UP000276834 UP000286641 UP000011712 UP000261480 UP000245340 UP000007754 UP000286640 UP000007635 UP000261620 UP000009046 UP000028760 UP000261500 UP000261380 UP000197619 UP000257160 UP000007755 UP000001075 UP000002356 UP000261680 UP000291021 UP000286642 UP000261640 UP000246464 UP000016665 UP000000437 UP000002281 UP000009136 UP000002852 UP000248482 UP000192224 UP000000715 UP000007648 UP000261400 UP000092124
UP000027135 UP000245037 UP000018467 UP000192220 UP000245341 UP000265080 UP000264840 UP000265100 UP000265160 UP000257200 UP000261460 UP000242638 UP000001555 UP000261600 UP000005207 UP000264800 UP000269221 UP000261340 UP000008912 UP000261440 UP000276834 UP000286641 UP000011712 UP000261480 UP000245340 UP000007754 UP000286640 UP000007635 UP000261620 UP000009046 UP000028760 UP000261500 UP000261380 UP000197619 UP000257160 UP000007755 UP000001075 UP000002356 UP000261680 UP000291021 UP000286642 UP000261640 UP000246464 UP000016665 UP000000437 UP000002281 UP000009136 UP000002852 UP000248482 UP000192224 UP000000715 UP000007648 UP000261400 UP000092124
Interpro
ProteinModelPortal
H9JGA7
A0A212ELA2
A0A194QM96
A0A1E1WAI0
A0A1E1WG20
A0A194RIH7
+ More
A0A3S2LY56 A0A0L7LAH9 A0A067RLA5 A0A0V0GCJ9 A0A2P8YGF1 A0A3B1JXT4 A0A224YNK8 A0A023G7P1 T1JCQ7 A0A2I4B122 A0A131XJL8 A0A1A7X9A2 A0A2U3YX08 A0A3P8UAN4 A0A0C9SC69 A0A3Q3CCN1 A0A3P8R923 A0A3P9BQB3 A0A3Q1EY05 A0A3B4GTB3 A0A3P9NJL6 A0A3Q1EA08 B7Q3F6 A0A315VR18 A0A131YR72 A0A3Q3IW50 I3JUQ2 A0A3Q3G760 A0A3M0J414 A0A3Q0RI01 A0A0S7HKA3 G1LF86 A0A3B4CKU8 A0A3L8SR64 D2HRE0 A0A131XVZ4 A0A3Q7PFV5 M3WRB4 A0A3B3YJN8 A0A2U3W7L1 H0Z676 A0A3Q7TSF4 G3NLE2 G3NLE4 V5HWA4 Q2WBW2 A0A3N0YVD1 A0A3Q3VVV1 E0VLE6 A0A087YGY6 A0A3B3TUG2 A0A3B5LRB9 A0A218V7F6 A0A3Q1AR02 A0A1A8JSV9 F4X1J1 G3GVW8 A0A1A8F434 W5Q9V5 K9IRU0 A0A384DML2 A0A3Q7WX20 A0A3Q3NA22 A0A2U9CCB9 U3JYR3 L8I3R8 B8A4D5 A0A3Q2HH00 Q1LZC4 A0A3B5R240 A0A2Y9IVI3 A0A1W4Z4I0 A0A1A8BCG0 G9L2D3 F6Q6G1 G3X2I1 A0A3B5A2M4 A0A1A8V9R6 A0A1A6HEX0
A0A3S2LY56 A0A0L7LAH9 A0A067RLA5 A0A0V0GCJ9 A0A2P8YGF1 A0A3B1JXT4 A0A224YNK8 A0A023G7P1 T1JCQ7 A0A2I4B122 A0A131XJL8 A0A1A7X9A2 A0A2U3YX08 A0A3P8UAN4 A0A0C9SC69 A0A3Q3CCN1 A0A3P8R923 A0A3P9BQB3 A0A3Q1EY05 A0A3B4GTB3 A0A3P9NJL6 A0A3Q1EA08 B7Q3F6 A0A315VR18 A0A131YR72 A0A3Q3IW50 I3JUQ2 A0A3Q3G760 A0A3M0J414 A0A3Q0RI01 A0A0S7HKA3 G1LF86 A0A3B4CKU8 A0A3L8SR64 D2HRE0 A0A131XVZ4 A0A3Q7PFV5 M3WRB4 A0A3B3YJN8 A0A2U3W7L1 H0Z676 A0A3Q7TSF4 G3NLE2 G3NLE4 V5HWA4 Q2WBW2 A0A3N0YVD1 A0A3Q3VVV1 E0VLE6 A0A087YGY6 A0A3B3TUG2 A0A3B5LRB9 A0A218V7F6 A0A3Q1AR02 A0A1A8JSV9 F4X1J1 G3GVW8 A0A1A8F434 W5Q9V5 K9IRU0 A0A384DML2 A0A3Q7WX20 A0A3Q3NA22 A0A2U9CCB9 U3JYR3 L8I3R8 B8A4D5 A0A3Q2HH00 Q1LZC4 A0A3B5R240 A0A2Y9IVI3 A0A1W4Z4I0 A0A1A8BCG0 G9L2D3 F6Q6G1 G3X2I1 A0A3B5A2M4 A0A1A8V9R6 A0A1A6HEX0
PDB
5ZYT
E-value=8.30734e-36,
Score=375
Ontologies
GO
Topology
Subcellular location
Mitochondrion
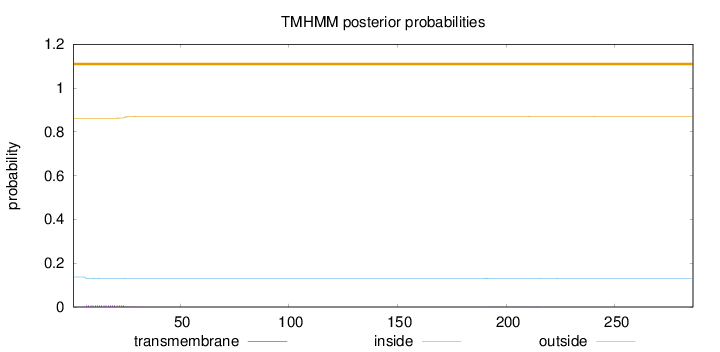
Length:
286
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.16961
Exp number, first 60 AAs:
0.14945
Total prob of N-in:
0.13751
outside
1 - 286
Population Genetic Test Statistics
Pi
236.087312
Theta
173.103833
Tajima's D
-1.259136
CLR
101.284193
CSRT
0.0918954052297385
Interpretation
Uncertain
