Gene
KWMTBOMO11067
Pre Gene Modal
BGIBMGA008308
Annotation
Connectin_[Papilio_machaon]
Location in the cell
Nuclear Reliability : 1.792
Sequence
CDS
ATGGAACAAGTCACCAAGTACCTGATCATCATAGCTTCCGTGATGACTGTTAAGCTTACTGTTACCGAAAGTAAGAATGATACCAAGCCCAGAATAGATAGAAGGTACTTTCACGTGCCCATGTTTGTTAACATCTGTGATATAGAAGACCGCAGATCCAAAGTGCATTGCTACTGCGATAACAACAAACCTAAAATGGCCACTCGAGCCGACTGCTGGGTGTTTGGAGGCGGAATAACTGAAGATGATCCAATATGGCCTAATTTTTCCTCACAAAGCGAATTGGAGCATCTTATCTTCAACGCTAGATCTGATGACGCTCTCACCTTTATTCCAGCGAAAGCCATTGTCAGATTAGATAAACTAAAACATGTAACCATACAGTTCGGTACTATCAAACGTATTCCACCGTACGCTTTCACAAATTCATCAACATTACGACAGATTGTATTGAAATCAACGAAGATAACGAAACTCGAGAAGCACGCTTTCTCGCGGCTGATGATGCTGTCAAACCTGAGTTTGGATGACAATCAAATAGAGGAATTGAAACGAGAATCGTTCTACAACCTACCGAATTTACACATACTCCACTTGTCGAACAATAACATTAGTTTGATACACGAAGGCTGCTTCAAACACCTGAACAACTTAATAGAATTAAGACTAGATTACAACTACATATCAGTAGTGACGCGGGAGATGTTCGAGGGACTGGAAAACCTCTCGAAATTGAACTTGAGACACAATAAGTTAACGATGATCGGAAATTTGGCGTTCACTGAACTGTGGGGCCTGAAAGAGCTGATGCTCGATAACAATGCTATAGAATACATATCGGAGAGAGCTTTTGGCGGTCTGGCTCAATTGAGAAAATTGTGGCTATCGGGGAATAGGTTGACGACATTTTACGAGGACGCTCTCGAAGATATGCGGAGTCTGGTAGTGTTGGATTTAAGAAATAATCTGTTGGAAACGATATCTTACGAGGCGATTCGACCACTGGTCGAGAATGTTAAATCAAGTTCGGCTGCAGTCTACTTAGAGGGTGAGTTGCAATACTCAAACTATAACTTATAA
Protein
MEQVTKYLIIIASVMTVKLTVTESKNDTKPRIDRRYFHVPMFVNICDIEDRRSKVHCYCDNNKPKMATRADCWVFGGGITEDDPIWPNFSSQSELEHLIFNARSDDALTFIPAKAIVRLDKLKHVTIQFGTIKRIPPYAFTNSSTLRQIVLKSTKITKLEKHAFSRLMMLSNLSLDDNQIEELKRESFYNLPNLHILHLSNNNISLIHEGCFKHLNNLIELRLDYNYISVVTREMFEGLENLSKLNLRHNKLTMIGNLAFTELWGLKELMLDNNAIEYISERAFGGLAQLRKLWLSGNRLTTFYEDALEDMRSLVVLDLRNNLLETISYEAIRPLVENVKSSSAAVYLEGELQYSNYNL
Summary
Uniprot
H9JFL1
A0A194QGQ1
A0A194RI53
A0A2H1X1V4
A0A2A4JNA6
A0A0L7LRI8
+ More
A0A3S2NX82 A0A212F0E9 A0A2H1VKA7 A0A0L7LJM4 H9JGE2 A0A2Z5U6Q8 A0A212EWB6 A0A194RIU1 A0A194QI49 A0A3S2LY46 A0A212FJY7 A0A0L7LT00 A0A2A4JHU6 A0A194RII6 A0A2H1WRJ5 H9JGE5 A0A194QGR0 A0A0L7LCW6 A0A2H1X0G1 A0A2A4IZZ4 A0A3S2NQN6 A0A212EY84 A0A0L7KSB1 A0A2H1WWK7 H9JGC9 A0A194RI80 A0A194QGJ2 A0A3S2P0I5 A0A194QMB6 A0A2J7RGY8 D1ZZZ7 A0A2P8YXU8 A0A2P9A985 A0A0L7QK14 A0A1B6EUX6 A0A1B6IKB2 A0A154PP99 E0VLF1 A0A1Y1KVT1 A0A1Y1KSF1 A0A2R7VVV6 A0A067RE87 A0A336L4T9 T1HG84 A0A088AP31 A0A1Y1MUS6 A0A1Y1MUR2 A0A182SV66 Q177Q1 A0A194RMP0 A0A084VHE4 E9I8W9 B0X9P1 A0A0M8ZTC3 A0A336LLJ4 A0A182HFA7 E2A8P1 A0A232EZM6 A0A1B6CMN5 A0A182NCF0 A0A182XBB6 A0A182KLR3 Q7PYM2 Q16QN1 A0A182U569 A0A182I059 A0A182IMQ6 A0A182VJ73 A0A0P4W4I6 A0A182MKY2 A0A182VZ11 A0A195C8C6 A0A182R6D0 A0A182QL42 A0A182PKK5 A0A182FKT3 W5JPC6 A0A182K4I7 A0A182XVW5 A0A182KH31 A0A1A9WLG2 A0A182XVW4 A0A195F1M2 A0A0P4W729 A0A1I8QD93 A0A0A9X110 A0A1A9ZUD0 A0A1A9YCJ8 A0A1B0B0B4 A0A182M1F9 B0X035 A0A182PKK4 A0A195EJP1 A0A182UD95
A0A3S2NX82 A0A212F0E9 A0A2H1VKA7 A0A0L7LJM4 H9JGE2 A0A2Z5U6Q8 A0A212EWB6 A0A194RIU1 A0A194QI49 A0A3S2LY46 A0A212FJY7 A0A0L7LT00 A0A2A4JHU6 A0A194RII6 A0A2H1WRJ5 H9JGE5 A0A194QGR0 A0A0L7LCW6 A0A2H1X0G1 A0A2A4IZZ4 A0A3S2NQN6 A0A212EY84 A0A0L7KSB1 A0A2H1WWK7 H9JGC9 A0A194RI80 A0A194QGJ2 A0A3S2P0I5 A0A194QMB6 A0A2J7RGY8 D1ZZZ7 A0A2P8YXU8 A0A2P9A985 A0A0L7QK14 A0A1B6EUX6 A0A1B6IKB2 A0A154PP99 E0VLF1 A0A1Y1KVT1 A0A1Y1KSF1 A0A2R7VVV6 A0A067RE87 A0A336L4T9 T1HG84 A0A088AP31 A0A1Y1MUS6 A0A1Y1MUR2 A0A182SV66 Q177Q1 A0A194RMP0 A0A084VHE4 E9I8W9 B0X9P1 A0A0M8ZTC3 A0A336LLJ4 A0A182HFA7 E2A8P1 A0A232EZM6 A0A1B6CMN5 A0A182NCF0 A0A182XBB6 A0A182KLR3 Q7PYM2 Q16QN1 A0A182U569 A0A182I059 A0A182IMQ6 A0A182VJ73 A0A0P4W4I6 A0A182MKY2 A0A182VZ11 A0A195C8C6 A0A182R6D0 A0A182QL42 A0A182PKK5 A0A182FKT3 W5JPC6 A0A182K4I7 A0A182XVW5 A0A182KH31 A0A1A9WLG2 A0A182XVW4 A0A195F1M2 A0A0P4W729 A0A1I8QD93 A0A0A9X110 A0A1A9ZUD0 A0A1A9YCJ8 A0A1B0B0B4 A0A182M1F9 B0X035 A0A182PKK4 A0A195EJP1 A0A182UD95
Pubmed
EMBL
BABH01002360
KQ458880
KPJ04594.1
KQ460154
KPJ17242.1
ODYU01012753
+ More
SOQ59218.1 NWSH01001048 PCG72923.1 JTDY01000246 KOB78072.1 RSAL01000035 RVE51318.1 AGBW02011096 OWR47219.1 ODYU01002867 SOQ40882.1 JTDY01000864 KOB75625.1 BABH01033551 AP017473 BBB06699.1 AGBW02012044 OWR45761.1 KPJ17255.1 KPJ04605.1 RSAL01000117 RVE46822.1 AGBW02008193 OWR54044.1 JTDY01000194 KOB78356.1 NWSH01001336 PCG71627.1 KPJ17254.1 ODYU01010456 SOQ55582.1 BABH01033535 KPJ04604.1 JTDY01001654 KOB73240.1 ODYU01012486 SOQ58819.1 NWSH01004345 PCG65199.1 RSAL01000372 RVE42134.1 AGBW02011598 OWR46445.1 JTDY01006497 KOB65961.1 ODYU01011209 SOQ56804.1 BABH01033645 KPJ17267.1 KPJ04617.1 RSAL01000068 RVE49260.1 KPJ04616.1 NEVH01003750 PNF40092.1 KQ971338 EFA02447.2 PYGN01000295 PSN49068.1 LT717637 SIW61629.1 KQ414990 KOC58968.1 GECZ01028051 GECZ01022424 GECZ01015237 GECZ01002416 JAS41718.1 JAS47345.1 JAS54532.1 JAS67353.1 GECU01020353 JAS87353.1 KQ435007 KZC13567.1 DS235271 EEB14207.1 GEZM01076966 GEZM01076965 GEZM01076964 GEZM01076963 JAV63506.1 GEZM01076967 GEZM01076962 JAV63508.1 KK854111 PTY11469.1 KK852559 KDR21353.1 UFQS01001597 UFQT01001597 SSX11539.1 SSX31106.1 ACPB03023550 GEZM01020065 GEZM01020063 JAV89424.1 GEZM01020064 JAV89423.1 CH477372 EAT42413.1 KPJ17266.1 ATLV01013162 KE524842 KFB37388.1 GL761662 EFZ22980.1 DS232543 EDS43243.1 KQ435859 KOX70547.1 UFQT01000045 SSX18826.1 JXUM01036703 JXUM01036704 JXUM01036705 KQ561103 KXJ79693.1 GL437657 EFN70198.1 NNAY01001478 OXU23825.1 GEDC01022564 GEDC01019421 GEDC01017721 JAS14734.1 JAS17877.1 JAS19577.1 AAAB01008987 EAA01440.5 CH477742 EAT36704.1 APCN01002046 GDRN01085066 JAI61404.1 AXCM01006762 KQ978081 KYM97087.1 AXCN02000019 ADMH02000940 ETN64619.1 KQ981856 KYN34475.1 GDRN01085074 JAI61403.1 GBHO01029960 JAG13644.1 JXJN01006686 AXCM01004687 DS232227 EDS37910.1 KQ978782 KYN28470.1
SOQ59218.1 NWSH01001048 PCG72923.1 JTDY01000246 KOB78072.1 RSAL01000035 RVE51318.1 AGBW02011096 OWR47219.1 ODYU01002867 SOQ40882.1 JTDY01000864 KOB75625.1 BABH01033551 AP017473 BBB06699.1 AGBW02012044 OWR45761.1 KPJ17255.1 KPJ04605.1 RSAL01000117 RVE46822.1 AGBW02008193 OWR54044.1 JTDY01000194 KOB78356.1 NWSH01001336 PCG71627.1 KPJ17254.1 ODYU01010456 SOQ55582.1 BABH01033535 KPJ04604.1 JTDY01001654 KOB73240.1 ODYU01012486 SOQ58819.1 NWSH01004345 PCG65199.1 RSAL01000372 RVE42134.1 AGBW02011598 OWR46445.1 JTDY01006497 KOB65961.1 ODYU01011209 SOQ56804.1 BABH01033645 KPJ17267.1 KPJ04617.1 RSAL01000068 RVE49260.1 KPJ04616.1 NEVH01003750 PNF40092.1 KQ971338 EFA02447.2 PYGN01000295 PSN49068.1 LT717637 SIW61629.1 KQ414990 KOC58968.1 GECZ01028051 GECZ01022424 GECZ01015237 GECZ01002416 JAS41718.1 JAS47345.1 JAS54532.1 JAS67353.1 GECU01020353 JAS87353.1 KQ435007 KZC13567.1 DS235271 EEB14207.1 GEZM01076966 GEZM01076965 GEZM01076964 GEZM01076963 JAV63506.1 GEZM01076967 GEZM01076962 JAV63508.1 KK854111 PTY11469.1 KK852559 KDR21353.1 UFQS01001597 UFQT01001597 SSX11539.1 SSX31106.1 ACPB03023550 GEZM01020065 GEZM01020063 JAV89424.1 GEZM01020064 JAV89423.1 CH477372 EAT42413.1 KPJ17266.1 ATLV01013162 KE524842 KFB37388.1 GL761662 EFZ22980.1 DS232543 EDS43243.1 KQ435859 KOX70547.1 UFQT01000045 SSX18826.1 JXUM01036703 JXUM01036704 JXUM01036705 KQ561103 KXJ79693.1 GL437657 EFN70198.1 NNAY01001478 OXU23825.1 GEDC01022564 GEDC01019421 GEDC01017721 JAS14734.1 JAS17877.1 JAS19577.1 AAAB01008987 EAA01440.5 CH477742 EAT36704.1 APCN01002046 GDRN01085066 JAI61404.1 AXCM01006762 KQ978081 KYM97087.1 AXCN02000019 ADMH02000940 ETN64619.1 KQ981856 KYN34475.1 GDRN01085074 JAI61403.1 GBHO01029960 JAG13644.1 JXJN01006686 AXCM01004687 DS232227 EDS37910.1 KQ978782 KYN28470.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000037510
UP000283053
+ More
UP000007151 UP000235965 UP000007266 UP000245037 UP000053825 UP000076502 UP000009046 UP000027135 UP000015103 UP000005203 UP000075901 UP000008820 UP000030765 UP000002320 UP000053105 UP000069940 UP000249989 UP000000311 UP000215335 UP000075884 UP000076407 UP000075882 UP000007062 UP000075902 UP000075840 UP000075880 UP000075903 UP000075883 UP000075920 UP000078542 UP000075900 UP000075886 UP000075885 UP000069272 UP000000673 UP000075881 UP000076408 UP000091820 UP000078541 UP000095300 UP000092445 UP000092443 UP000092460 UP000078492
UP000007151 UP000235965 UP000007266 UP000245037 UP000053825 UP000076502 UP000009046 UP000027135 UP000015103 UP000005203 UP000075901 UP000008820 UP000030765 UP000002320 UP000053105 UP000069940 UP000249989 UP000000311 UP000215335 UP000075884 UP000076407 UP000075882 UP000007062 UP000075902 UP000075840 UP000075880 UP000075903 UP000075883 UP000075920 UP000078542 UP000075900 UP000075886 UP000075885 UP000069272 UP000000673 UP000075881 UP000076408 UP000091820 UP000078541 UP000095300 UP000092445 UP000092443 UP000092460 UP000078492
Interpro
Gene 3D
ProteinModelPortal
H9JFL1
A0A194QGQ1
A0A194RI53
A0A2H1X1V4
A0A2A4JNA6
A0A0L7LRI8
+ More
A0A3S2NX82 A0A212F0E9 A0A2H1VKA7 A0A0L7LJM4 H9JGE2 A0A2Z5U6Q8 A0A212EWB6 A0A194RIU1 A0A194QI49 A0A3S2LY46 A0A212FJY7 A0A0L7LT00 A0A2A4JHU6 A0A194RII6 A0A2H1WRJ5 H9JGE5 A0A194QGR0 A0A0L7LCW6 A0A2H1X0G1 A0A2A4IZZ4 A0A3S2NQN6 A0A212EY84 A0A0L7KSB1 A0A2H1WWK7 H9JGC9 A0A194RI80 A0A194QGJ2 A0A3S2P0I5 A0A194QMB6 A0A2J7RGY8 D1ZZZ7 A0A2P8YXU8 A0A2P9A985 A0A0L7QK14 A0A1B6EUX6 A0A1B6IKB2 A0A154PP99 E0VLF1 A0A1Y1KVT1 A0A1Y1KSF1 A0A2R7VVV6 A0A067RE87 A0A336L4T9 T1HG84 A0A088AP31 A0A1Y1MUS6 A0A1Y1MUR2 A0A182SV66 Q177Q1 A0A194RMP0 A0A084VHE4 E9I8W9 B0X9P1 A0A0M8ZTC3 A0A336LLJ4 A0A182HFA7 E2A8P1 A0A232EZM6 A0A1B6CMN5 A0A182NCF0 A0A182XBB6 A0A182KLR3 Q7PYM2 Q16QN1 A0A182U569 A0A182I059 A0A182IMQ6 A0A182VJ73 A0A0P4W4I6 A0A182MKY2 A0A182VZ11 A0A195C8C6 A0A182R6D0 A0A182QL42 A0A182PKK5 A0A182FKT3 W5JPC6 A0A182K4I7 A0A182XVW5 A0A182KH31 A0A1A9WLG2 A0A182XVW4 A0A195F1M2 A0A0P4W729 A0A1I8QD93 A0A0A9X110 A0A1A9ZUD0 A0A1A9YCJ8 A0A1B0B0B4 A0A182M1F9 B0X035 A0A182PKK4 A0A195EJP1 A0A182UD95
A0A3S2NX82 A0A212F0E9 A0A2H1VKA7 A0A0L7LJM4 H9JGE2 A0A2Z5U6Q8 A0A212EWB6 A0A194RIU1 A0A194QI49 A0A3S2LY46 A0A212FJY7 A0A0L7LT00 A0A2A4JHU6 A0A194RII6 A0A2H1WRJ5 H9JGE5 A0A194QGR0 A0A0L7LCW6 A0A2H1X0G1 A0A2A4IZZ4 A0A3S2NQN6 A0A212EY84 A0A0L7KSB1 A0A2H1WWK7 H9JGC9 A0A194RI80 A0A194QGJ2 A0A3S2P0I5 A0A194QMB6 A0A2J7RGY8 D1ZZZ7 A0A2P8YXU8 A0A2P9A985 A0A0L7QK14 A0A1B6EUX6 A0A1B6IKB2 A0A154PP99 E0VLF1 A0A1Y1KVT1 A0A1Y1KSF1 A0A2R7VVV6 A0A067RE87 A0A336L4T9 T1HG84 A0A088AP31 A0A1Y1MUS6 A0A1Y1MUR2 A0A182SV66 Q177Q1 A0A194RMP0 A0A084VHE4 E9I8W9 B0X9P1 A0A0M8ZTC3 A0A336LLJ4 A0A182HFA7 E2A8P1 A0A232EZM6 A0A1B6CMN5 A0A182NCF0 A0A182XBB6 A0A182KLR3 Q7PYM2 Q16QN1 A0A182U569 A0A182I059 A0A182IMQ6 A0A182VJ73 A0A0P4W4I6 A0A182MKY2 A0A182VZ11 A0A195C8C6 A0A182R6D0 A0A182QL42 A0A182PKK5 A0A182FKT3 W5JPC6 A0A182K4I7 A0A182XVW5 A0A182KH31 A0A1A9WLG2 A0A182XVW4 A0A195F1M2 A0A0P4W729 A0A1I8QD93 A0A0A9X110 A0A1A9ZUD0 A0A1A9YCJ8 A0A1B0B0B4 A0A182M1F9 B0X035 A0A182PKK4 A0A195EJP1 A0A182UD95
PDB
4QXF
E-value=9.02951e-18,
Score=221
Ontologies
GO
Topology
SignalP
Position: 1 - 24,
Likelihood: 0.602034
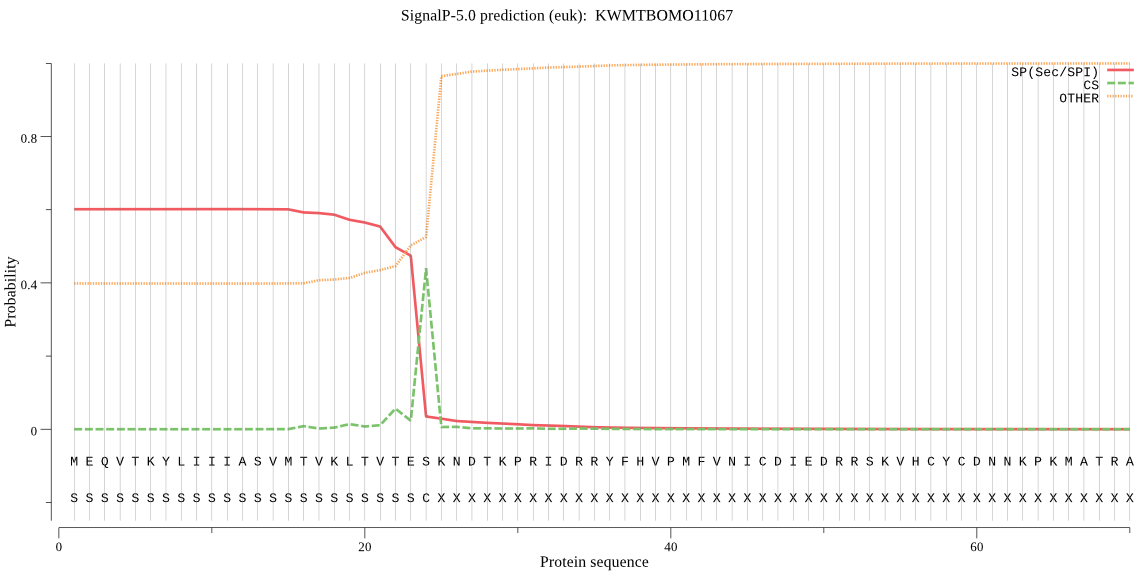
Length:
359
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.30465
Exp number, first 60 AAs:
0.30351
Total prob of N-in:
0.03483
outside
1 - 359

Population Genetic Test Statistics
Pi
239.339792
Theta
196.083271
Tajima's D
0.691296
CLR
0
CSRT
0.567521623918804
Interpretation
Uncertain
