Gene
KWMTBOMO11064 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008551
Annotation
PREDICTED:_hypoxia_up-regulated_protein_1_[Papilio_polytes]
Location in the cell
Cytoplasmic Reliability : 1.735 ER Reliability : 1.356
Sequence
CDS
ATGTATAGTGTGTTGTGTGTGGTGTTATTGCCGATACTTTTAAGCCAACAAGTCGATGCGGCGGCAGTAATTTCCATTGATTTTGGATCAGAATGGATGAAGGTTGGCATAGTGTCACCGGGTGTTCCTATGGAAATCGTGTTGAACAAGGAGTCTAAGAGGAAGACACCAGCTGTCGTTGCCTTCAGAGATGATGTTCGGACATTCGGGGAAGATGCTGTGACAGTTGGAGTCAGATTCCCTAAAAATTCCTACAAATATTTACTTGAACTTCTTGGCAAGACATATGACCACCCTCTTGTACAATTATTCAGAAAGAGGTTCCCTTATTATGAAATTGTGGAATCGGACCGAGGCACAATTGAATTTGTGCATGATGACAAGACAAAATTTACACCTGAAGAGCTAGTTGCACAGATTCTAGCCAAAGCCAAAGATTTTGCTGAAATTAATCATGGCCAACAGATTAGCGAGTGTGTGATTACAGTTCCTGGTTACTTTAATCAGGCTGAGCGTAGAGCACTCAAAGAAGCTGCTTCATTAGCAGGCTTGAATGTTTTGCAGTTAATTAATGATTATACTGCTATTGCTATCAACTATGGCATATTTAGGAGAAAGGAAATCAATGAAACCGCATGGTACACTTTGTTCTTTGATATGGGAGCATCTTCGACTAAAGCCGCATTAGTTGAATATAAAACTGTCAAAATTAAGGACAAGGGTTATGTTGAAACTGTGCCACAACTGCAGGTCTTAGGTGTTGGTTTTGACAGAACACTTGGAGGTTTAGAAATGACGCTTCGTCTCCGCGAGCATCTGATTAAAGCCTGGGAGGCTAACGGGGGAGGCGATGTGCGAGCGTCACCTCGTGCTATGGAGAAACTGTTCCGCGAGGCGGAAAGGCTAAAGGTCGTGCTCTCTGCTAACACCGAGTTCTACGCTCAAATTGAAAGCCTTCTTGATGACAAAGACTTCAAGCTGCAGGTGAGCAGGGCTGAGCTGGAAGAGTTATGTGCTGACCTCTGGGCGCGGGTGGGGGCGGTGGTGCGGCGCGCGGTGTCGGCGGCGGGGGGCGCGGGGGCGGGCGCGCGGCTGGTGGTGGCGGGGGGCGGGTCGCGCGTGCCCGCCGTGCTGGCCGCGCTCGCCGACGCGGTGCAGCACGAGCCCGCGCGCTCCATCAACGCGGACGAGGCTGCCACTCTCGGAGCTGTCTACAGAGCGGCATCATTAGCAACAGGATACAAAGTGGCCCCTCTGAATGTACGAGATGCTGTGTTGCTTCCAATACAAGTTGTCTTCACAAGACACGCCGATGGCGTCGACAAGCTGGTTAAGAGAACTCTCTTTGGGCCTATGAACCCCTACCCACAGAAGAAGGTGATCACTTTCAATAAGCACACAGAGGATTTCTCCTTCAACGTGAACTATGCAGAACTTGATCACATCCCATCCACTGAATTGCAGTACATTGGTGCTCTGAACTTGAGTGAGGTTTCGTTGAGTGGAGTTTCCGCGGCTTTGGGTAAACATGTTGGCGAAAATGTCGAACATAAAGGTATTAAGGCACATTTCAATATGGACGACTCCGGCATACTAAACTTGGTTAACGTTGAATTTGTTGCTGAGAAGACCGTCACAGAAGACGAAGACAAAGATAGCACTCTGTCTAAGATCGGAAGTACTATTAGTAAATTATTTGGTTCAGATTCAGAAACTCCGGAAAAAACTGAAGAACAACCCCAGGGAGAAGCTAAGAACGATACACAGAATCTCAACGAGACGACCACTGATAAGCAGAATTCAACAGAAACTGACAGCAAACCTAAACCGAAAATCGTTATTTTGAAAGAACCAATTAAATCCGAAGAATTAGTCCTCAACTTGCAGCCTCTGTCAACCGACCAGTTCAAGAATTCGAAAGCAAAGATAAATGAACTAAACACGATCGACAAGAAGCGTGTCCAAAGAGAAACGGCGTTGAATAATCTCGAAGCGTTCGTTGTTGACGCCCAAATGAAGATCGACATGGAGGAGTATGCTGAATGTGGAACTGAGGAGGAAATTGAGGAGATCCGTAAACTGTGCAGCGAGACTTCCGAATGGTTATACGATGATGGATACGATGCAGACACTGAACTGTTCGAACAAAAACTTAATGCTCTTAAAGAAAAAACTCATCCCATATTCTACAAGCATTGGGAACACAGAGAAAGGGCGGACGCGGTTGCAGCGATTCGAAACTTATTGAATAGTTCTAAGGAATTCCTTAAAATGTCAAAGAACTTCACCAAAGAGTCGAATGAAGAGAAAGATGTCTTTACCGAAGTTGAGATTGGCGTTTTAGAAAAGAAGATCGAGGAAACTGAGACTTGGCTAGAGAAGGCCATCAAAGAACAAAAGGCACTGAAAAAGAACGAAGACGTTAAATTGACAGTTGACAGCATTCGAGAAAAAATGGCAAATCTCGACAGAGAAGTAAAGTATTTGTTGAACAAAATGAAGATTTGGCGACCCAAGAAACCAGTCAAGAAAGATGCAGATAATAAAACCGAGGAAGTCGTGATAGAGCCTCCAGAGAATCCCGAGACTCAGGAGGAGGCTCCTCAAGATGATAGCGGTGCTGATGATAAACCAGAAGAAGAGGTTAAAGAGATGGACGCTAAAGAAGAATCAACAGAGCCACCCCTACAGCTCTCTGACGGTCAAGCTGAACATTCCGAGTTATAA
Protein
MYSVLCVVLLPILLSQQVDAAAVISIDFGSEWMKVGIVSPGVPMEIVLNKESKRKTPAVVAFRDDVRTFGEDAVTVGVRFPKNSYKYLLELLGKTYDHPLVQLFRKRFPYYEIVESDRGTIEFVHDDKTKFTPEELVAQILAKAKDFAEINHGQQISECVITVPGYFNQAERRALKEAASLAGLNVLQLINDYTAIAINYGIFRRKEINETAWYTLFFDMGASSTKAALVEYKTVKIKDKGYVETVPQLQVLGVGFDRTLGGLEMTLRLREHLIKAWEANGGGDVRASPRAMEKLFREAERLKVVLSANTEFYAQIESLLDDKDFKLQVSRAELEELCADLWARVGAVVRRAVSAAGGAGAGARLVVAGGGSRVPAVLAALADAVQHEPARSINADEAATLGAVYRAASLATGYKVAPLNVRDAVLLPIQVVFTRHADGVDKLVKRTLFGPMNPYPQKKVITFNKHTEDFSFNVNYAELDHIPSTELQYIGALNLSEVSLSGVSAALGKHVGENVEHKGIKAHFNMDDSGILNLVNVEFVAEKTVTEDEDKDSTLSKIGSTISKLFGSDSETPEKTEEQPQGEAKNDTQNLNETTTDKQNSTETDSKPKPKIVILKEPIKSEELVLNLQPLSTDQFKNSKAKINELNTIDKKRVQRETALNNLEAFVVDAQMKIDMEEYAECGTEEEIEEIRKLCSETSEWLYDDGYDADTELFEQKLNALKEKTHPIFYKHWEHRERADAVAAIRNLLNSSKEFLKMSKNFTKESNEEKDVFTEVEIGVLEKKIEETETWLEKAIKEQKALKKNEDVKLTVDSIREKMANLDREVKYLLNKMKIWRPKKPVKKDADNKTEEVVIEPPENPETQEEAPQDDSGADDKPEEEVKEMDAKEESTEPPLQLSDGQAEHSEL
Summary
Similarity
Belongs to the heat shock protein 70 family.
Uniprot
H9JGA4
A0A1L6UW57
A0A3S2NMB7
A0A2A4J222
A0A2H1WD30
A0A194PGL6
+ More
A0A212FA98 S4NXF8 A0A194RIS5 A0A1W4WWZ7 A0A1Y1M3G5 A0A0T6BHH0 D2A1Y3 N6TRK0 U4UKB7 A0A067RL26 J9HG73 A0A1S4G5N6 A0A1L8DU22 U5EW42 A0A2J7PUI5 A0A2J7PUJ5 A0A023EWK4 A0A182PR32 A0A182Q0R7 A0A1W7R800 A0A182K425 A0A182JHS1 A0A232FLN9 A0A182YJX8 A0A182N4Y7 A0A1Q3FAB9 A0A1Q3FAL8 K7INW6 A0A182SXE1 A0A1Q3FA24 A0A2M3YYX7 A0A1Q3F9W9 A0A2M3YYQ4 A0A224XE47 A0A0N0BIV3 A0A182MPB6 B0WKR8 A0A182F6X2 A0A2M4BCA9 A0A084VRQ9 A0A411G6N6 A0A2M3YYW3 A0A2M3ZZN5 Q7PYB0 A0A154P7S5 A0A2M3ZZS3 A0A0K8TS50 A0A182I0I9 A0A2M3ZZM4 A0A0L7RI19 A0A182RBY4 A0A182X2N8 A0A182UQ00 A0A146LN42 W5JLW0 A0A1I9WL37 A0A2M4CGB0 A0A0C9QZD6 A0A0P4VS14 V9ILC7 A0A088ARD8 A0A2K8JV05 R4WQM8 A0A182VQU5 A0A0A1XB50 A0A1B6CKK6 A0A1W6EWB4 B4L5V3 A0A1S4N1I5 E0W3D7 W8C075 E2C858 A0A336K1X8 A0A336LJS3 B4M7E2 A0A182U555 A0A034VS83 A0A0K8UUM5 B4R3H2 A0A0M4EUQ0 A0A0C9RPQ7 O46067 B4I9T7 A0A1W4UPT9 B4Q175 F4W487 B3P902 A0A0Q5T728 A0A195E5K9 A0A195ET77 A0A1J1HJA8 A0A026VV58 A0A195CJL8 A0A1I8PTD8
A0A212FA98 S4NXF8 A0A194RIS5 A0A1W4WWZ7 A0A1Y1M3G5 A0A0T6BHH0 D2A1Y3 N6TRK0 U4UKB7 A0A067RL26 J9HG73 A0A1S4G5N6 A0A1L8DU22 U5EW42 A0A2J7PUI5 A0A2J7PUJ5 A0A023EWK4 A0A182PR32 A0A182Q0R7 A0A1W7R800 A0A182K425 A0A182JHS1 A0A232FLN9 A0A182YJX8 A0A182N4Y7 A0A1Q3FAB9 A0A1Q3FAL8 K7INW6 A0A182SXE1 A0A1Q3FA24 A0A2M3YYX7 A0A1Q3F9W9 A0A2M3YYQ4 A0A224XE47 A0A0N0BIV3 A0A182MPB6 B0WKR8 A0A182F6X2 A0A2M4BCA9 A0A084VRQ9 A0A411G6N6 A0A2M3YYW3 A0A2M3ZZN5 Q7PYB0 A0A154P7S5 A0A2M3ZZS3 A0A0K8TS50 A0A182I0I9 A0A2M3ZZM4 A0A0L7RI19 A0A182RBY4 A0A182X2N8 A0A182UQ00 A0A146LN42 W5JLW0 A0A1I9WL37 A0A2M4CGB0 A0A0C9QZD6 A0A0P4VS14 V9ILC7 A0A088ARD8 A0A2K8JV05 R4WQM8 A0A182VQU5 A0A0A1XB50 A0A1B6CKK6 A0A1W6EWB4 B4L5V3 A0A1S4N1I5 E0W3D7 W8C075 E2C858 A0A336K1X8 A0A336LJS3 B4M7E2 A0A182U555 A0A034VS83 A0A0K8UUM5 B4R3H2 A0A0M4EUQ0 A0A0C9RPQ7 O46067 B4I9T7 A0A1W4UPT9 B4Q175 F4W487 B3P902 A0A0Q5T728 A0A195E5K9 A0A195ET77 A0A1J1HJA8 A0A026VV58 A0A195CJL8 A0A1I8PTD8
Pubmed
19121390
26354079
22118469
23622113
28004739
18362917
+ More
19820115 23537049 24845553 17510324 24945155 28648823 25244985 20075255 24438588 12364791 14747013 17210077 26369729 26823975 20920257 23761445 27538518 27129103 23691247 25830018 17994087 20566863 24495485 20798317 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 21719571 24508170 30249741
19820115 23537049 24845553 17510324 24945155 28648823 25244985 20075255 24438588 12364791 14747013 17210077 26369729 26823975 20920257 23761445 27538518 27129103 23691247 25830018 17994087 20566863 24495485 20798317 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 21719571 24508170 30249741
EMBL
BABH01002326
KY379955
APS85765.1
RSAL01000035
RVE51320.1
NWSH01003698
+ More
PCG65909.1 ODYU01007832 SOQ50937.1 KQ459604 KPI92447.1 AGBW02009508 OWR50650.1 GAIX01012192 JAA80368.1 KQ460154 KPJ17240.1 GEZM01042808 JAV79578.1 LJIG01000319 KRT86621.1 KQ971338 EFA02143.1 APGK01022393 APGK01022394 KB740420 ENN80698.1 KB632390 ERL94524.1 KK852450 KDR23698.1 CH477636 EJY57847.1 GFDF01004158 JAV09926.1 GANO01000723 JAB59148.1 NEVH01021197 PNF19992.1 PNF19993.1 GAPW01000140 JAC13458.1 AXCN02000185 GEHC01000379 JAV47266.1 NNAY01000039 OXU31666.1 GFDL01010545 JAV24500.1 GFDL01010429 JAV24616.1 GFDL01010611 JAV24434.1 GGFM01000667 MBW21418.1 GFDL01010674 JAV24371.1 GGFM01000653 MBW21404.1 GFTR01008358 JAW08068.1 KQ435726 KOX78218.1 AXCM01005541 DS231975 EDS30010.1 GGFJ01001516 MBW50657.1 ATLV01015768 KE525036 KFB40653.1 MH365729 QBB01538.1 GGFM01000695 MBW21446.1 GGFK01000692 MBW34013.1 AAAB01008987 EAA01085.3 KQ434825 KZC07374.1 GGFK01000678 MBW33999.1 GDAI01000424 JAI17179.1 APCN01002086 GGFK01000676 MBW33997.1 KQ414584 KOC70637.1 GDHC01010024 JAQ08605.1 ADMH02001196 ETN63764.1 KU932221 APA33857.1 GGFL01000202 MBW64380.1 GBYB01009159 GBYB01009163 JAG78926.1 JAG78930.1 GDKW01001229 JAI55366.1 JR050456 AEY61286.1 KY031144 ATU82895.1 AK417986 BAN21201.1 GBXI01006282 JAD08010.1 GEDC01027149 GEDC01023413 GEDC01018485 GEDC01006718 GEDC01003827 JAS10149.1 JAS13885.1 JAS18813.1 JAS30580.1 JAS33471.1 KY563607 ARK20016.1 CH933811 EDW06562.1 AAZO01007370 DS235882 EEB20143.1 GAMC01007544 JAB99011.1 GL453568 EFN75861.1 UFQS01000021 UFQT01000021 SSW97518.1 SSX17904.1 SSW97519.1 SSX17905.1 CH940653 EDW62709.1 GAKP01012796 JAC46156.1 GDHF01022259 GDHF01007732 JAI30055.1 JAI44582.1 CM000366 EDX16924.1 CP012528 ALC49526.1 GBYB01009161 JAG78928.1 AE014298 AY094705 AL009196 AAF45769.1 AAM11058.1 ACZ95182.1 CAA15711.1 CH480825 EDW43968.1 CM000162 EDX01382.1 KRK06132.1 KRK06133.1 KRK06134.1 GL887491 EGI71083.1 CH954183 EDV45607.2 KQS26036.1 KQ979609 KYN20182.1 KQ981979 KYN31455.1 CVRI01000006 CRK88111.1 KK107796 QOIP01000013 EZA47658.1 RLU15578.1 KQ977642 KYN00925.1
PCG65909.1 ODYU01007832 SOQ50937.1 KQ459604 KPI92447.1 AGBW02009508 OWR50650.1 GAIX01012192 JAA80368.1 KQ460154 KPJ17240.1 GEZM01042808 JAV79578.1 LJIG01000319 KRT86621.1 KQ971338 EFA02143.1 APGK01022393 APGK01022394 KB740420 ENN80698.1 KB632390 ERL94524.1 KK852450 KDR23698.1 CH477636 EJY57847.1 GFDF01004158 JAV09926.1 GANO01000723 JAB59148.1 NEVH01021197 PNF19992.1 PNF19993.1 GAPW01000140 JAC13458.1 AXCN02000185 GEHC01000379 JAV47266.1 NNAY01000039 OXU31666.1 GFDL01010545 JAV24500.1 GFDL01010429 JAV24616.1 GFDL01010611 JAV24434.1 GGFM01000667 MBW21418.1 GFDL01010674 JAV24371.1 GGFM01000653 MBW21404.1 GFTR01008358 JAW08068.1 KQ435726 KOX78218.1 AXCM01005541 DS231975 EDS30010.1 GGFJ01001516 MBW50657.1 ATLV01015768 KE525036 KFB40653.1 MH365729 QBB01538.1 GGFM01000695 MBW21446.1 GGFK01000692 MBW34013.1 AAAB01008987 EAA01085.3 KQ434825 KZC07374.1 GGFK01000678 MBW33999.1 GDAI01000424 JAI17179.1 APCN01002086 GGFK01000676 MBW33997.1 KQ414584 KOC70637.1 GDHC01010024 JAQ08605.1 ADMH02001196 ETN63764.1 KU932221 APA33857.1 GGFL01000202 MBW64380.1 GBYB01009159 GBYB01009163 JAG78926.1 JAG78930.1 GDKW01001229 JAI55366.1 JR050456 AEY61286.1 KY031144 ATU82895.1 AK417986 BAN21201.1 GBXI01006282 JAD08010.1 GEDC01027149 GEDC01023413 GEDC01018485 GEDC01006718 GEDC01003827 JAS10149.1 JAS13885.1 JAS18813.1 JAS30580.1 JAS33471.1 KY563607 ARK20016.1 CH933811 EDW06562.1 AAZO01007370 DS235882 EEB20143.1 GAMC01007544 JAB99011.1 GL453568 EFN75861.1 UFQS01000021 UFQT01000021 SSW97518.1 SSX17904.1 SSW97519.1 SSX17905.1 CH940653 EDW62709.1 GAKP01012796 JAC46156.1 GDHF01022259 GDHF01007732 JAI30055.1 JAI44582.1 CM000366 EDX16924.1 CP012528 ALC49526.1 GBYB01009161 JAG78928.1 AE014298 AY094705 AL009196 AAF45769.1 AAM11058.1 ACZ95182.1 CAA15711.1 CH480825 EDW43968.1 CM000162 EDX01382.1 KRK06132.1 KRK06133.1 KRK06134.1 GL887491 EGI71083.1 CH954183 EDV45607.2 KQS26036.1 KQ979609 KYN20182.1 KQ981979 KYN31455.1 CVRI01000006 CRK88111.1 KK107796 QOIP01000013 EZA47658.1 RLU15578.1 KQ977642 KYN00925.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000007151
UP000053240
+ More
UP000192223 UP000007266 UP000019118 UP000030742 UP000027135 UP000008820 UP000235965 UP000075885 UP000075886 UP000075881 UP000075880 UP000215335 UP000076408 UP000075884 UP000002358 UP000075901 UP000053105 UP000075883 UP000002320 UP000069272 UP000030765 UP000007062 UP000076502 UP000075840 UP000053825 UP000075900 UP000076407 UP000075903 UP000000673 UP000005203 UP000075920 UP000009192 UP000009046 UP000008237 UP000008792 UP000075902 UP000000304 UP000092553 UP000000803 UP000001292 UP000192221 UP000002282 UP000007755 UP000008711 UP000078492 UP000078541 UP000183832 UP000053097 UP000279307 UP000078542 UP000095300
UP000192223 UP000007266 UP000019118 UP000030742 UP000027135 UP000008820 UP000235965 UP000075885 UP000075886 UP000075881 UP000075880 UP000215335 UP000076408 UP000075884 UP000002358 UP000075901 UP000053105 UP000075883 UP000002320 UP000069272 UP000030765 UP000007062 UP000076502 UP000075840 UP000053825 UP000075900 UP000076407 UP000075903 UP000000673 UP000005203 UP000075920 UP000009192 UP000009046 UP000008237 UP000008792 UP000075902 UP000000304 UP000092553 UP000000803 UP000001292 UP000192221 UP000002282 UP000007755 UP000008711 UP000078492 UP000078541 UP000183832 UP000053097 UP000279307 UP000078542 UP000095300
Interpro
Gene 3D
ProteinModelPortal
H9JGA4
A0A1L6UW57
A0A3S2NMB7
A0A2A4J222
A0A2H1WD30
A0A194PGL6
+ More
A0A212FA98 S4NXF8 A0A194RIS5 A0A1W4WWZ7 A0A1Y1M3G5 A0A0T6BHH0 D2A1Y3 N6TRK0 U4UKB7 A0A067RL26 J9HG73 A0A1S4G5N6 A0A1L8DU22 U5EW42 A0A2J7PUI5 A0A2J7PUJ5 A0A023EWK4 A0A182PR32 A0A182Q0R7 A0A1W7R800 A0A182K425 A0A182JHS1 A0A232FLN9 A0A182YJX8 A0A182N4Y7 A0A1Q3FAB9 A0A1Q3FAL8 K7INW6 A0A182SXE1 A0A1Q3FA24 A0A2M3YYX7 A0A1Q3F9W9 A0A2M3YYQ4 A0A224XE47 A0A0N0BIV3 A0A182MPB6 B0WKR8 A0A182F6X2 A0A2M4BCA9 A0A084VRQ9 A0A411G6N6 A0A2M3YYW3 A0A2M3ZZN5 Q7PYB0 A0A154P7S5 A0A2M3ZZS3 A0A0K8TS50 A0A182I0I9 A0A2M3ZZM4 A0A0L7RI19 A0A182RBY4 A0A182X2N8 A0A182UQ00 A0A146LN42 W5JLW0 A0A1I9WL37 A0A2M4CGB0 A0A0C9QZD6 A0A0P4VS14 V9ILC7 A0A088ARD8 A0A2K8JV05 R4WQM8 A0A182VQU5 A0A0A1XB50 A0A1B6CKK6 A0A1W6EWB4 B4L5V3 A0A1S4N1I5 E0W3D7 W8C075 E2C858 A0A336K1X8 A0A336LJS3 B4M7E2 A0A182U555 A0A034VS83 A0A0K8UUM5 B4R3H2 A0A0M4EUQ0 A0A0C9RPQ7 O46067 B4I9T7 A0A1W4UPT9 B4Q175 F4W487 B3P902 A0A0Q5T728 A0A195E5K9 A0A195ET77 A0A1J1HJA8 A0A026VV58 A0A195CJL8 A0A1I8PTD8
A0A212FA98 S4NXF8 A0A194RIS5 A0A1W4WWZ7 A0A1Y1M3G5 A0A0T6BHH0 D2A1Y3 N6TRK0 U4UKB7 A0A067RL26 J9HG73 A0A1S4G5N6 A0A1L8DU22 U5EW42 A0A2J7PUI5 A0A2J7PUJ5 A0A023EWK4 A0A182PR32 A0A182Q0R7 A0A1W7R800 A0A182K425 A0A182JHS1 A0A232FLN9 A0A182YJX8 A0A182N4Y7 A0A1Q3FAB9 A0A1Q3FAL8 K7INW6 A0A182SXE1 A0A1Q3FA24 A0A2M3YYX7 A0A1Q3F9W9 A0A2M3YYQ4 A0A224XE47 A0A0N0BIV3 A0A182MPB6 B0WKR8 A0A182F6X2 A0A2M4BCA9 A0A084VRQ9 A0A411G6N6 A0A2M3YYW3 A0A2M3ZZN5 Q7PYB0 A0A154P7S5 A0A2M3ZZS3 A0A0K8TS50 A0A182I0I9 A0A2M3ZZM4 A0A0L7RI19 A0A182RBY4 A0A182X2N8 A0A182UQ00 A0A146LN42 W5JLW0 A0A1I9WL37 A0A2M4CGB0 A0A0C9QZD6 A0A0P4VS14 V9ILC7 A0A088ARD8 A0A2K8JV05 R4WQM8 A0A182VQU5 A0A0A1XB50 A0A1B6CKK6 A0A1W6EWB4 B4L5V3 A0A1S4N1I5 E0W3D7 W8C075 E2C858 A0A336K1X8 A0A336LJS3 B4M7E2 A0A182U555 A0A034VS83 A0A0K8UUM5 B4R3H2 A0A0M4EUQ0 A0A0C9RPQ7 O46067 B4I9T7 A0A1W4UPT9 B4Q175 F4W487 B3P902 A0A0Q5T728 A0A195E5K9 A0A195ET77 A0A1J1HJA8 A0A026VV58 A0A195CJL8 A0A1I8PTD8
PDB
3JXU
E-value=3.13658e-42,
Score=436
Ontologies
GO
Topology
SignalP
Position: 1 - 20,
Likelihood: 0.998330
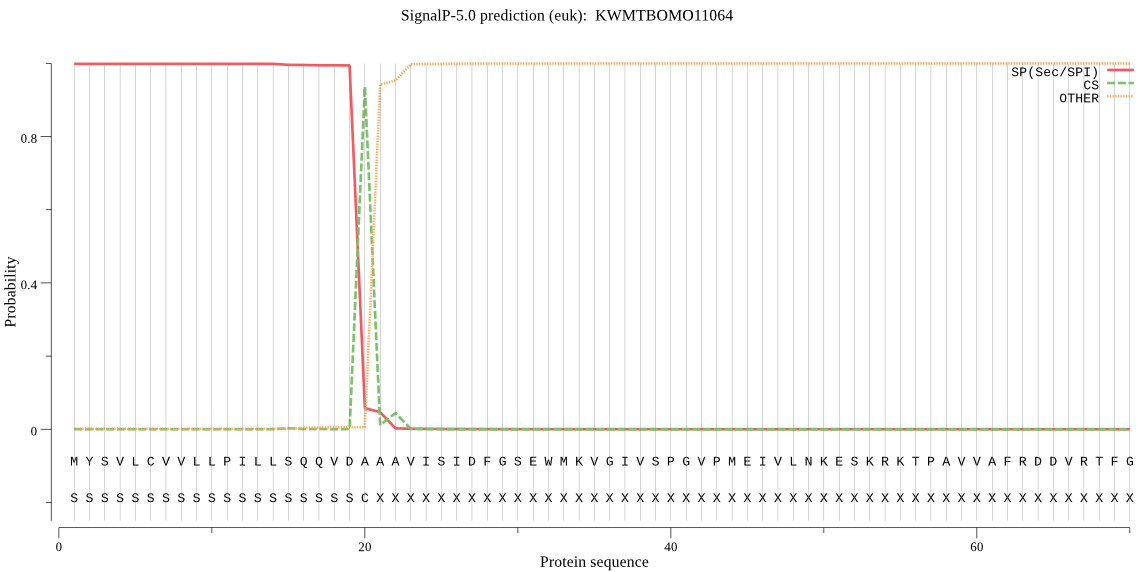
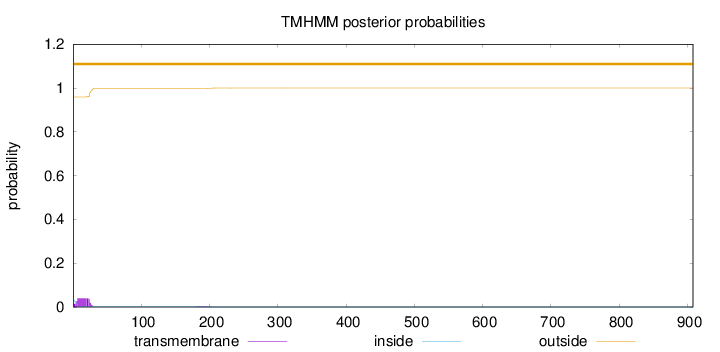
Length:
908
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.899729999999999
Exp number, first 60 AAs:
0.86727
Total prob of N-in:
0.04069
outside
1 - 908
Population Genetic Test Statistics
Pi
213.964572
Theta
169.194017
Tajima's D
0.536425
CLR
0.781154
CSRT
0.526823658817059
Interpretation
Uncertain
