Gene
KWMTBOMO11061 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008312
Annotation
mitochondrial_prohibitin_complex_protein_2_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.88
Sequence
CDS
ATGGCACAAAGTAAGATAAATGACATGGCAGGAAAATTCGCTAAAGGTGGGCCACCTGGTCTTGGCATCGGTCTCAAAGTGGTAGCCGTTGTCGGCGCTGCGGCTTATGGCGTCTCCCAATCGGTGTTTACCGTGGAAGGAGGTCATCGTGCCATTATGTTCAACAGAATAGGAGGAGTACAACAGCACGTATTCACTGAAGGTATGCACTTCCGTATACCGTGGTTTCAATACCCCATCATTTATGATATCAGATCCCGACCTCGCAAGATATCTTCCCCGACTGGTTCAAAAGATTTACAGATGGTAAACATTTCCCTCCGAGTACTTTCTCGACCAGATGCCAATATGCTAGCAACAATGTACAGACAACTTGGAACAGATTATGATGAAAAGGTTTTGCCATCTATATGCAATGAAGTCCTAAAATCGGTTGTTGCCAAATTCAATGCCTCCCAGCTGATCACACAGCGTCAGCAGGTGTCTCTACTAATCAGAAGAGAGTTGGTTGAGCGTGCTGCTGATTTTAATATTATCCTTGATGATGTATCCCTCACTGAACTAAGCTTTGGTAAGGAATACACAGCTGCAGTAGAAGCTAAACAAGTTGCACAACAGGAGGCTCAGCGTGCAGCTTTTGTAGTTGAAAGGGCAAAGCAAGAGCGTCAACAGAAAATTGTGCAAGCAGAGGGAGAAGCTGAAGCTGCTGAGATGTTGGGTAAAGCCATGGGCATGAACCCTGGTTATTTAAAACTTAGGAAAATTCGTGCTGCTCAAAGTATTTCGAGAATGATTGCTCAATCTCAGAACCGTGTCTTCCTCCCTGGAAACAGTCTGATGATCAATCTTCAAGATCCTACTTTTGATGATTTGTCTGAGAAACTCACTAAAAAGAAGTAA
Protein
MAQSKINDMAGKFAKGGPPGLGIGLKVVAVVGAAAYGVSQSVFTVEGGHRAIMFNRIGGVQQHVFTEGMHFRIPWFQYPIIYDIRSRPRKISSPTGSKDLQMVNISLRVLSRPDANMLATMYRQLGTDYDEKVLPSICNEVLKSVVAKFNASQLITQRQQVSLLIRRELVERAADFNIILDDVSLTELSFGKEYTAAVEAKQVAQQEAQRAAFVVERAKQERQQKIVQAEGEAEAAEMLGKAMGMNPGYLKLRKIRAAQSISRMIAQSQNRVFLPGNSLMINLQDPTFDDLSEKLTKKK
Summary
Uniprot
Q2F5N1
H6T598
A0A2A4K017
I4DKA3
A0A212EJV7
A0A194RI48
+ More
A0A0L7LH04 A0A2H1WFV1 A0A2J7RFG0 A0A2J7RFF9 A0A067QGU8 A0A1Y1M0C5 A0A182SIM8 A0A0T6B5F9 A0A182JYK2 A0A084WFV4 A0A182NAZ1 A0A023EQ60 A0A182J2W9 A0A2M4AAF1 A0A182QG56 A0A023EMU2 A0A1W4W553 A0A2M4CNN1 A0A182Y8B9 A0A182F5X8 Q7PMG2 A0A182LVF6 A0A182UA48 A0A182ICR7 A0A0A9YIU9 A0A2M4CPQ6 A0A182GGR9 A0A182PKU0 Q16MJ0 A0A0A9YPT1 A0A182VA57 T1HZB4 A0A1W4W573 A0A0A9YKB4 A0A182VTV5 A0A224XPB6 A0A0N7Z9D5 R4G5G9 A0A2M3Z4R7 A0A2M4A9V6 A0A2M4BVG4 Q16HQ0 J3JYC4 R4WDC2 A0A139WG95 A0A1Q3F744 E0VDG8 A0A069DRL9 Q16MJ2 A0A170Z2N9 A0A182L654 A0A023FAC5 A0A1B6D3F7 D2A682 W5JB53 A0A1J1IQ62 A0A0V0G4A2 A0A182WZF0 V5GXD7 B0W1R7 A0A240SY15 A0A182RGP3 A0A0K8TSS7 A0A1B6CTA3 U5EXV4 A0A1Q3G247 K0FIK6 A0A034WBC4 A0A1L8DU16 W8C189 A0A1B6EPW9 A0A336LXX3 N6TRG1 A0A1L8DU15 A0A1B6IZQ6 A0A151X2A0 K0FEE3 A2I473 A0A0K8VBK0 A0A1B0DL95 A0A0A1X9J6 A0A0K8UYK9 A0A336M1W5 A0A1B6FUB2 A0A1B6GYM6 A0A1B6GK12 A0A0K8WDG2 A0A0A1WZ21 A0A1B6INL6 A0A151IE80 A0A3B0IZT0 A0A1I8ML31 A0A195BYU4 A0A158NM74
A0A0L7LH04 A0A2H1WFV1 A0A2J7RFG0 A0A2J7RFF9 A0A067QGU8 A0A1Y1M0C5 A0A182SIM8 A0A0T6B5F9 A0A182JYK2 A0A084WFV4 A0A182NAZ1 A0A023EQ60 A0A182J2W9 A0A2M4AAF1 A0A182QG56 A0A023EMU2 A0A1W4W553 A0A2M4CNN1 A0A182Y8B9 A0A182F5X8 Q7PMG2 A0A182LVF6 A0A182UA48 A0A182ICR7 A0A0A9YIU9 A0A2M4CPQ6 A0A182GGR9 A0A182PKU0 Q16MJ0 A0A0A9YPT1 A0A182VA57 T1HZB4 A0A1W4W573 A0A0A9YKB4 A0A182VTV5 A0A224XPB6 A0A0N7Z9D5 R4G5G9 A0A2M3Z4R7 A0A2M4A9V6 A0A2M4BVG4 Q16HQ0 J3JYC4 R4WDC2 A0A139WG95 A0A1Q3F744 E0VDG8 A0A069DRL9 Q16MJ2 A0A170Z2N9 A0A182L654 A0A023FAC5 A0A1B6D3F7 D2A682 W5JB53 A0A1J1IQ62 A0A0V0G4A2 A0A182WZF0 V5GXD7 B0W1R7 A0A240SY15 A0A182RGP3 A0A0K8TSS7 A0A1B6CTA3 U5EXV4 A0A1Q3G247 K0FIK6 A0A034WBC4 A0A1L8DU16 W8C189 A0A1B6EPW9 A0A336LXX3 N6TRG1 A0A1L8DU15 A0A1B6IZQ6 A0A151X2A0 K0FEE3 A2I473 A0A0K8VBK0 A0A1B0DL95 A0A0A1X9J6 A0A0K8UYK9 A0A336M1W5 A0A1B6FUB2 A0A1B6GYM6 A0A1B6GK12 A0A0K8WDG2 A0A0A1WZ21 A0A1B6INL6 A0A151IE80 A0A3B0IZT0 A0A1I8ML31 A0A195BYU4 A0A158NM74
Pubmed
19121390
22651552
26354079
22118469
26227816
24845553
+ More
28004739 24438588 24945155 26483478 25244985 12364791 25401762 26823975 17510324 27129103 22516182 23691247 18362917 19820115 20566863 26334808 20966253 25474469 20920257 23761445 26369729 25348373 24495485 23537049 25830018 25315136 21347285
28004739 24438588 24945155 26483478 25244985 12364791 25401762 26823975 17510324 27129103 22516182 23691247 18362917 19820115 20566863 26334808 20966253 25474469 20920257 23761445 26369729 25348373 24495485 23537049 25830018 25315136 21347285
EMBL
BABH01002322
DQ311392
ABD36336.1
HQ337624
ADQ90002.1
NWSH01000359
+ More
PCG77010.1 AK401721 KQ459604 BAM18343.1 KPI92450.1 AGBW02014369 OWR41779.1 KQ460154 KPJ17237.1 JTDY01001108 KOB74853.1 ODYU01008394 SOQ51960.1 NEVH01004413 PNF39567.1 PNF39568.1 KK853474 KDR07361.1 GEZM01045565 GEZM01045564 JAV77745.1 LJIG01009712 KRT82532.1 ATLV01023406 KE525343 KFB49098.1 JXUM01152402 GAPW01002914 KQ571446 JAC10684.1 KXJ68175.1 GGFK01004389 MBW37710.1 AXCN02000651 GAPW01002921 JAC10677.1 GGFL01002691 MBW66869.1 AAAB01008980 EAA13889.4 AXCM01001667 APCN01005755 GBHO01012067 GBHO01012063 GBRD01010596 GBRD01010594 GDHC01014664 JAG31537.1 JAG31541.1 JAG55228.1 JAQ03965.1 GGFL01002690 MBW66868.1 JXUM01062272 JXUM01062273 KQ562192 KXJ76445.1 CH477860 EAT35551.1 GBHO01012064 GBRD01010595 GBRD01010592 GDHC01011072 JAG31540.1 JAG55229.1 JAQ07557.1 ACPB03025237 GBHO01012066 GBRD01010593 GDHC01017467 JAG31538.1 JAG55231.1 JAQ01162.1 GFTR01004748 JAW11678.1 GDKW01000851 JAI55744.1 GAHY01000580 JAA76930.1 GGFM01002753 MBW23504.1 GGFK01004265 MBW37586.1 GGFJ01007872 MBW57013.1 CH478151 EAT33777.1 EAT35550.1 BT128250 AEE63210.1 AK417424 BAN20639.1 KQ971346 KYB26993.1 GFDL01011655 JAV23390.1 DS235076 EEB11424.1 GBGD01002319 JAC86570.1 EAT35549.1 GEMB01002672 JAS00521.1 GBBI01000773 JAC17939.1 GEDC01017066 JAS20232.1 EFA05476.1 ADMH02001884 ETN60638.1 CVRI01000057 CRL02359.1 GECL01003259 JAP02865.1 GALX01003498 JAB64968.1 DS231823 EDS26618.1 AJWK01006928 GDAI01000422 JAI17181.1 GEDC01020529 JAS16769.1 GANO01000732 JAB59139.1 GFDL01001160 JAV33885.1 JX317625 AFU08568.1 GAKP01006953 JAC51999.1 GFDF01004145 JAV09939.1 GAMC01007095 GAMC01007094 GAMC01007093 GAMC01007092 JAB99461.1 GECZ01029784 JAS39985.1 UFQT01000285 UFQT01000393 SSX22824.1 SSX23945.1 APGK01007420 APGK01007424 KB737275 KB737274 KB632191 ENN83049.1 ENN83055.1 ERL89831.1 GFDF01004187 JAV09897.1 GECU01015314 JAS92392.1 KQ982584 KYQ54408.1 JX317624 AFU08567.1 EF070576 ABM55642.1 GDHF01016394 JAI35920.1 AJVK01070774 GBXI01006328 JAD07964.1 GDHF01020585 JAI31729.1 SSX22823.1 GECZ01016196 JAS53573.1 GECZ01002254 JAS67515.1 GECZ01007011 JAS62758.1 GDHF01003225 JAI49089.1 GBXI01010391 JAD03901.1 GECU01019191 JAS88515.1 KQ977890 KYM98982.1 OUUW01000001 SPP73677.1 KQ976395 KYM93096.1 ADTU01020332
PCG77010.1 AK401721 KQ459604 BAM18343.1 KPI92450.1 AGBW02014369 OWR41779.1 KQ460154 KPJ17237.1 JTDY01001108 KOB74853.1 ODYU01008394 SOQ51960.1 NEVH01004413 PNF39567.1 PNF39568.1 KK853474 KDR07361.1 GEZM01045565 GEZM01045564 JAV77745.1 LJIG01009712 KRT82532.1 ATLV01023406 KE525343 KFB49098.1 JXUM01152402 GAPW01002914 KQ571446 JAC10684.1 KXJ68175.1 GGFK01004389 MBW37710.1 AXCN02000651 GAPW01002921 JAC10677.1 GGFL01002691 MBW66869.1 AAAB01008980 EAA13889.4 AXCM01001667 APCN01005755 GBHO01012067 GBHO01012063 GBRD01010596 GBRD01010594 GDHC01014664 JAG31537.1 JAG31541.1 JAG55228.1 JAQ03965.1 GGFL01002690 MBW66868.1 JXUM01062272 JXUM01062273 KQ562192 KXJ76445.1 CH477860 EAT35551.1 GBHO01012064 GBRD01010595 GBRD01010592 GDHC01011072 JAG31540.1 JAG55229.1 JAQ07557.1 ACPB03025237 GBHO01012066 GBRD01010593 GDHC01017467 JAG31538.1 JAG55231.1 JAQ01162.1 GFTR01004748 JAW11678.1 GDKW01000851 JAI55744.1 GAHY01000580 JAA76930.1 GGFM01002753 MBW23504.1 GGFK01004265 MBW37586.1 GGFJ01007872 MBW57013.1 CH478151 EAT33777.1 EAT35550.1 BT128250 AEE63210.1 AK417424 BAN20639.1 KQ971346 KYB26993.1 GFDL01011655 JAV23390.1 DS235076 EEB11424.1 GBGD01002319 JAC86570.1 EAT35549.1 GEMB01002672 JAS00521.1 GBBI01000773 JAC17939.1 GEDC01017066 JAS20232.1 EFA05476.1 ADMH02001884 ETN60638.1 CVRI01000057 CRL02359.1 GECL01003259 JAP02865.1 GALX01003498 JAB64968.1 DS231823 EDS26618.1 AJWK01006928 GDAI01000422 JAI17181.1 GEDC01020529 JAS16769.1 GANO01000732 JAB59139.1 GFDL01001160 JAV33885.1 JX317625 AFU08568.1 GAKP01006953 JAC51999.1 GFDF01004145 JAV09939.1 GAMC01007095 GAMC01007094 GAMC01007093 GAMC01007092 JAB99461.1 GECZ01029784 JAS39985.1 UFQT01000285 UFQT01000393 SSX22824.1 SSX23945.1 APGK01007420 APGK01007424 KB737275 KB737274 KB632191 ENN83049.1 ENN83055.1 ERL89831.1 GFDF01004187 JAV09897.1 GECU01015314 JAS92392.1 KQ982584 KYQ54408.1 JX317624 AFU08567.1 EF070576 ABM55642.1 GDHF01016394 JAI35920.1 AJVK01070774 GBXI01006328 JAD07964.1 GDHF01020585 JAI31729.1 SSX22823.1 GECZ01016196 JAS53573.1 GECZ01002254 JAS67515.1 GECZ01007011 JAS62758.1 GDHF01003225 JAI49089.1 GBXI01010391 JAD03901.1 GECU01019191 JAS88515.1 KQ977890 KYM98982.1 OUUW01000001 SPP73677.1 KQ976395 KYM93096.1 ADTU01020332
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000037510
+ More
UP000235965 UP000027135 UP000075901 UP000075881 UP000030765 UP000075884 UP000069940 UP000249989 UP000075880 UP000075886 UP000192223 UP000076408 UP000069272 UP000007062 UP000075883 UP000075902 UP000075840 UP000075885 UP000008820 UP000075903 UP000015103 UP000075920 UP000007266 UP000009046 UP000075882 UP000000673 UP000183832 UP000076407 UP000002320 UP000092461 UP000075900 UP000019118 UP000030742 UP000075809 UP000092462 UP000078542 UP000268350 UP000095301 UP000078540 UP000005205
UP000235965 UP000027135 UP000075901 UP000075881 UP000030765 UP000075884 UP000069940 UP000249989 UP000075880 UP000075886 UP000192223 UP000076408 UP000069272 UP000007062 UP000075883 UP000075902 UP000075840 UP000075885 UP000008820 UP000075903 UP000015103 UP000075920 UP000007266 UP000009046 UP000075882 UP000000673 UP000183832 UP000076407 UP000002320 UP000092461 UP000075900 UP000019118 UP000030742 UP000075809 UP000092462 UP000078542 UP000268350 UP000095301 UP000078540 UP000005205
Pfam
PF01145 Band_7
SUPFAM
SSF117892
SSF117892
ProteinModelPortal
Q2F5N1
H6T598
A0A2A4K017
I4DKA3
A0A212EJV7
A0A194RI48
+ More
A0A0L7LH04 A0A2H1WFV1 A0A2J7RFG0 A0A2J7RFF9 A0A067QGU8 A0A1Y1M0C5 A0A182SIM8 A0A0T6B5F9 A0A182JYK2 A0A084WFV4 A0A182NAZ1 A0A023EQ60 A0A182J2W9 A0A2M4AAF1 A0A182QG56 A0A023EMU2 A0A1W4W553 A0A2M4CNN1 A0A182Y8B9 A0A182F5X8 Q7PMG2 A0A182LVF6 A0A182UA48 A0A182ICR7 A0A0A9YIU9 A0A2M4CPQ6 A0A182GGR9 A0A182PKU0 Q16MJ0 A0A0A9YPT1 A0A182VA57 T1HZB4 A0A1W4W573 A0A0A9YKB4 A0A182VTV5 A0A224XPB6 A0A0N7Z9D5 R4G5G9 A0A2M3Z4R7 A0A2M4A9V6 A0A2M4BVG4 Q16HQ0 J3JYC4 R4WDC2 A0A139WG95 A0A1Q3F744 E0VDG8 A0A069DRL9 Q16MJ2 A0A170Z2N9 A0A182L654 A0A023FAC5 A0A1B6D3F7 D2A682 W5JB53 A0A1J1IQ62 A0A0V0G4A2 A0A182WZF0 V5GXD7 B0W1R7 A0A240SY15 A0A182RGP3 A0A0K8TSS7 A0A1B6CTA3 U5EXV4 A0A1Q3G247 K0FIK6 A0A034WBC4 A0A1L8DU16 W8C189 A0A1B6EPW9 A0A336LXX3 N6TRG1 A0A1L8DU15 A0A1B6IZQ6 A0A151X2A0 K0FEE3 A2I473 A0A0K8VBK0 A0A1B0DL95 A0A0A1X9J6 A0A0K8UYK9 A0A336M1W5 A0A1B6FUB2 A0A1B6GYM6 A0A1B6GK12 A0A0K8WDG2 A0A0A1WZ21 A0A1B6INL6 A0A151IE80 A0A3B0IZT0 A0A1I8ML31 A0A195BYU4 A0A158NM74
A0A0L7LH04 A0A2H1WFV1 A0A2J7RFG0 A0A2J7RFF9 A0A067QGU8 A0A1Y1M0C5 A0A182SIM8 A0A0T6B5F9 A0A182JYK2 A0A084WFV4 A0A182NAZ1 A0A023EQ60 A0A182J2W9 A0A2M4AAF1 A0A182QG56 A0A023EMU2 A0A1W4W553 A0A2M4CNN1 A0A182Y8B9 A0A182F5X8 Q7PMG2 A0A182LVF6 A0A182UA48 A0A182ICR7 A0A0A9YIU9 A0A2M4CPQ6 A0A182GGR9 A0A182PKU0 Q16MJ0 A0A0A9YPT1 A0A182VA57 T1HZB4 A0A1W4W573 A0A0A9YKB4 A0A182VTV5 A0A224XPB6 A0A0N7Z9D5 R4G5G9 A0A2M3Z4R7 A0A2M4A9V6 A0A2M4BVG4 Q16HQ0 J3JYC4 R4WDC2 A0A139WG95 A0A1Q3F744 E0VDG8 A0A069DRL9 Q16MJ2 A0A170Z2N9 A0A182L654 A0A023FAC5 A0A1B6D3F7 D2A682 W5JB53 A0A1J1IQ62 A0A0V0G4A2 A0A182WZF0 V5GXD7 B0W1R7 A0A240SY15 A0A182RGP3 A0A0K8TSS7 A0A1B6CTA3 U5EXV4 A0A1Q3G247 K0FIK6 A0A034WBC4 A0A1L8DU16 W8C189 A0A1B6EPW9 A0A336LXX3 N6TRG1 A0A1L8DU15 A0A1B6IZQ6 A0A151X2A0 K0FEE3 A2I473 A0A0K8VBK0 A0A1B0DL95 A0A0A1X9J6 A0A0K8UYK9 A0A336M1W5 A0A1B6FUB2 A0A1B6GYM6 A0A1B6GK12 A0A0K8WDG2 A0A0A1WZ21 A0A1B6INL6 A0A151IE80 A0A3B0IZT0 A0A1I8ML31 A0A195BYU4 A0A158NM74
Ontologies
GO
PANTHER
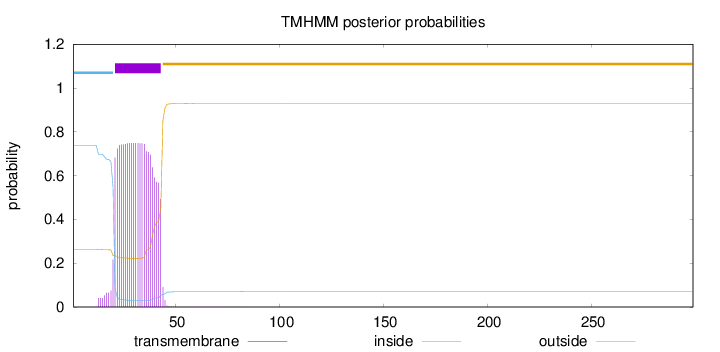
Topology
Length:
299
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
16.83716
Exp number, first 60 AAs:
16.81672
Total prob of N-in:
0.73765
POSSIBLE N-term signal
sequence
inside
1 - 20
TMhelix
21 - 43
outside
44 - 299
Population Genetic Test Statistics
Pi
125.159447
Theta
119.149467
Tajima's D
0.0202
CLR
0.10635
CSRT
0.370131493425329
Interpretation
Uncertain
