Gene
KWMTBOMO11058
Pre Gene Modal
BGIBMGA008315
Annotation
PREDICTED:_DNA_repair_protein_REV1_[Bombyx_mori]
Full name
DNA repair protein REV1
Location in the cell
Nuclear Reliability : 2.742
Sequence
CDS
ATGAGTCGAAATAAACGTGATGATAATTTCCCCGACAATGGGTTCGAGGCATGGGGAGGCTATATGCAAGCCAAAATTGCGAAACTGGAAAATCAGTTTACTTCAGTTACCGCAAAAGAAGAAATTCTTTCGAATATTTTCACTGGTATTAATATATTTGTAAATGGATTTACTATACCTTCAGCAGATGAACTTAAGTGTTTAATGTCCAAACACGGCGGTGTTTATCACACTTATCAAAGAAACGAAGATTTCATTATCGCCACAAATTTGCCAGATACGAAAGTGAAGAATATGTCTTTAGCGAGGGTGGTTAAGCCTGAATGGATAACTGATAGCATTAAAGCGAAAAAGTTACTAGATTACAGGAATTACTTGCTTTTTGGACATTCAAGAACACAAAGAGTGTTAAACTTCACAAAGAGAGTGACAAATGACAAAGATATAGATGAAACGAAATATAGTTCACTCAAAGAAACTTTAATTGAAAATAAAGATACAATAGTAAACAACGATTATTTCAATTCTATAAATTTAAAAAAAAGTACAATAAATACAACAAAACAAATGTGCCAAAACAGTATTAATGGAACTGGAAATGAGATAAATCCGGAAAAGTTGCAGGTCACAAAAACTGCAGCAGATCCCAACTTTATTTCTGAGTTCTACAACAACTCAAGGCTACATCATATTTCACAATTGGGTGCATGTTTCAAGCAGTATGTGAATGATTTGAGAGAACAAAGTAATTTTGTATTTCCCGCAAGACAAAATCTGAAGGACATAATTAAATCATCAAATAATCGATTATCGCAATTAGTCTTTGAAAAAGGAAAAACAATAATGCATATTGATATGGATTGTTTTTTTGTGTCAGTTGGGCTACGTAATAGGCCAGAGTTGAGAGGTAAACCTGTAGCTGTGACGCATTCTAAAGGAGGAATACCGCACATCAAAAGATCTGGTGTAGACAGGAATGCAGAATTTAATTTGTATAAACAGAAACAGGCCAAAAAACTTGTGAAAGACACTGGGTTACAGGGTCAAGAATTGGAAATTGAAACTAGAGTTGATAATATTGGAGATGAGGAGAGTAAATTTGGGTCCATGAGTGAGATAGCATCATGTTCATATGAAGCTAGGGCCAGGGGAGTTGGTAATGGTATGTTTATGGGTGTAGCGTTAAGATTGTGTCCTGATTTACAAACCATTCCATATGATTTTGAGGGTTACAAAGAAGTGGCTTATACTTTATACAATACTGTAGCCCAGTATACATTAGACATAGAAGCAGTCTCATGTGATGAGATGTATGTTGACTGTACAGAACTATTGAAAGATATGAACATGAATGTGCAGGATTTTGCTACAATTTTGAGAGAAGAGATTAAAACAAAAACTGGATGTCCTTGTTCAACAGGATTTGGAGGAAATCGGTTACAAGCAAGGTTGGCCACCAAAAAAGCTAAGCCTGATGGCCAGTTCTTTCTAACTGCAGATCTGGTACAGGATTTCATGTATGAAATCAATCTCAAAGACTTACCAGGTGTCGGTAGACAAATATCACAGAAATTGGAGTTATTGGGGCATACAACTTGTGGAACATTACAGAGTTTATCTTTAGCCACATTAAAATGCCATTTGGGCACTAAAACTGGAGCACAACTGTTTGATCAATGCAGAGGTCGTGGCCAAAATGCTTTAGCATTTCACACTGTGCGAAAATCAGTTTCTGCTGAAGTTAACTATGGTATTAGATTCGAAAATAATGATCAAGCTATTGAATTTTTGAAACAGTTGTCTGCTGAGGTACATTCAAGAATGCAGCAATTTAAAGTTTTAGGAAAATGTATAACTTTGAAGTTGATGGTCAGGGCTGAAGAAGCACCAGTTGAGACTGCAAAGTTTATGGGTCATGGATATTGTAATGTCATTAATAAATCAAATAATCTACCTGCAGCTACTAATGATGTTGAAGTGCTAACCCAGGAAACTATTGAGATATTTAAAAAAATAAATGTAGATGCTAAAGAAATGAGAGGTGTTGGAATACAAGTAACAAAATTGGTGCTAATTATTAAACAGCAAAGTAAAGGTGCCATCAGTAAATTTCTTTGTAACAAACCAACAATCATAAATATCAAAAGAGAATCCAAAACTGAAGTTGTTTCAAACAAAATAAACTTAGAGAACAATGAAAATAAACAACTTCAATTGAAAAATTCTAGGCTAAAGAAAAAATCTCCAACAACTGATAAAACTAAATCACCACCAGGGAAAAGACGAGGAAGACCACCCAAGGCACTTACTTTAGAACAAAAAGAAAAAAAACGTGCCATAAGTAAATCCGGAGTAGAAGCCAGAGATACACCATTCAAGAATCCAATGATGAAACCACAAAATTCACTCAAAGAAACAAAAATTAAAGTTGAAAAAACGGAAGGCCTCTTAGGTCTGCCGTGGAATAAAGCGAGAGAATTACTGAGAGCTTGGTTAGACAGTGGCCAAAAACCTCTAAGTTGCGATATCAGAATGATAGCAGAGTATATGAGAGAGATGGTAATTAAAAAAGAAATTGACAAAGTCGAAATATTGATAAACTTTTTAAGCAGAAGGACTGAGGAAATTGGTAGTCAAGTCTGGCTGGATGTGTATTGCCATGTTGTCGAACAAGTCCAGAATGCTATGATAGCTATTTACGGGACAAGGTTACATATTGACGCAACCTGA
Protein
MSRNKRDDNFPDNGFEAWGGYMQAKIAKLENQFTSVTAKEEILSNIFTGINIFVNGFTIPSADELKCLMSKHGGVYHTYQRNEDFIIATNLPDTKVKNMSLARVVKPEWITDSIKAKKLLDYRNYLLFGHSRTQRVLNFTKRVTNDKDIDETKYSSLKETLIENKDTIVNNDYFNSINLKKSTINTTKQMCQNSINGTGNEINPEKLQVTKTAADPNFISEFYNNSRLHHISQLGACFKQYVNDLREQSNFVFPARQNLKDIIKSSNNRLSQLVFEKGKTIMHIDMDCFFVSVGLRNRPELRGKPVAVTHSKGGIPHIKRSGVDRNAEFNLYKQKQAKKLVKDTGLQGQELEIETRVDNIGDEESKFGSMSEIASCSYEARARGVGNGMFMGVALRLCPDLQTIPYDFEGYKEVAYTLYNTVAQYTLDIEAVSCDEMYVDCTELLKDMNMNVQDFATILREEIKTKTGCPCSTGFGGNRLQARLATKKAKPDGQFFLTADLVQDFMYEINLKDLPGVGRQISQKLELLGHTTCGTLQSLSLATLKCHLGTKTGAQLFDQCRGRGQNALAFHTVRKSVSAEVNYGIRFENNDQAIEFLKQLSAEVHSRMQQFKVLGKCITLKLMVRAEEAPVETAKFMGHGYCNVINKSNNLPAATNDVEVLTQETIEIFKKINVDAKEMRGVGIQVTKLVLIIKQQSKGAISKFLCNKPTIINIKRESKTEVVSNKINLENNENKQLQLKNSRLKKKSPTTDKTKSPPGKRRGRPPKALTLEQKEKKRAISKSGVEARDTPFKNPMMKPQNSLKETKIKVEKTEGLLGLPWNKARELLRAWLDSGQKPLSCDIRMIAEYMREMVIKKEIDKVEILINFLSRRTEEIGSQVWLDVYCHVVEQVQNAMIAIYGTRLHIDAT
Summary
Description
Deoxycytidyl transferase involved in DNA repair. Transfers a dCMP residue from dCTP to the 3'-end of a DNA primer in a template-dependent reaction. May assist in the first step in the bypass of abasic lesions by the insertion of a nucleotide opposite the lesion. Required for normal induction of mutations by physical and chemical agents.
Similarity
Belongs to the DNA polymerase type-Y family.
Belongs to the mitochondrial carrier (TC 2.A.29) family.
Belongs to the mitochondrial carrier (TC 2.A.29) family.
Feature
chain DNA repair protein REV1
Uniprot
H9JFL8
A0A2H1WZM8
A0A0L7LTL5
A0A2A4JYB7
A0A212EJX8
A0A194PIA3
+ More
A0A067R8Z4 A0A1B6D5V3 A0A0A1WDT0 A0A0L0C0Z9 A0A1I8ND87 A0A2Z5TZG2 A0A1A9WC86 A0A1A9ZMM3 A0A1I8PNV4 A0A3Q0JLE4 A0A023F1M3 A0A034V8J5 A0A0K8VX46 A0A0K8VYB8 A0A0K8VCJ4 A0A1Q3EYU1 A0A1A9XPD7 A0A1B0AQ86 B0WKB9 A0A151WLK4 A0A1B0FJR5 A0A1A9V359 A0A195BNY4 A0A1W4W2L6 A0A0L7R6C0 T1HD53 E9INC1 A0A182FCB0 E1ZYK2 N6TT88 U4URC7 A0A232EXZ3 F4WJ40 A0A0M4E8N8 A0A182JB10 A0A151JRB0 A0A182PV68 A0A084WA92 A0A182MNF0 A0A0P4VTD3 A0A182YD29 Q7Q6B2 B4LGK8 Q29DW0 B4MLB6 A0A182KPP2 A0A182WKR2 A0A336KCQ3 A0A3R7MPY2 A0A0P8XTL6 B3M507 Q9W0P2 A0A3L8DCS6 A0A182QM37 A9UNE7 A0A026W820 A0A182VH23 A0A182JNL6 A0A0J7KS11 B4J2S6 A0A182X700 B4PCL6 A0A182NIP1 B4L0A6 B3NJA3 A0A088ASQ5 A0A0J9RLC9 A0A182I1Y4 A0A182UC77 A0A0A9X5V3 A0A0K8SS66 A0A3B0JZI7 E9H1D8 A0A0P6GJG4 A0A0P5F857 E0VNC0 A0A0P5SKK5 A0A0P5HQN3 T1J4C2 A0A2P8Z5H3 A0A151NXA6 A0A151NX22 A0A2T7PUI3 R7U747 A0A1W3JQL8 E9QIK8
A0A067R8Z4 A0A1B6D5V3 A0A0A1WDT0 A0A0L0C0Z9 A0A1I8ND87 A0A2Z5TZG2 A0A1A9WC86 A0A1A9ZMM3 A0A1I8PNV4 A0A3Q0JLE4 A0A023F1M3 A0A034V8J5 A0A0K8VX46 A0A0K8VYB8 A0A0K8VCJ4 A0A1Q3EYU1 A0A1A9XPD7 A0A1B0AQ86 B0WKB9 A0A151WLK4 A0A1B0FJR5 A0A1A9V359 A0A195BNY4 A0A1W4W2L6 A0A0L7R6C0 T1HD53 E9INC1 A0A182FCB0 E1ZYK2 N6TT88 U4URC7 A0A232EXZ3 F4WJ40 A0A0M4E8N8 A0A182JB10 A0A151JRB0 A0A182PV68 A0A084WA92 A0A182MNF0 A0A0P4VTD3 A0A182YD29 Q7Q6B2 B4LGK8 Q29DW0 B4MLB6 A0A182KPP2 A0A182WKR2 A0A336KCQ3 A0A3R7MPY2 A0A0P8XTL6 B3M507 Q9W0P2 A0A3L8DCS6 A0A182QM37 A9UNE7 A0A026W820 A0A182VH23 A0A182JNL6 A0A0J7KS11 B4J2S6 A0A182X700 B4PCL6 A0A182NIP1 B4L0A6 B3NJA3 A0A088ASQ5 A0A0J9RLC9 A0A182I1Y4 A0A182UC77 A0A0A9X5V3 A0A0K8SS66 A0A3B0JZI7 E9H1D8 A0A0P6GJG4 A0A0P5F857 E0VNC0 A0A0P5SKK5 A0A0P5HQN3 T1J4C2 A0A2P8Z5H3 A0A151NXA6 A0A151NX22 A0A2T7PUI3 R7U747 A0A1W3JQL8 E9QIK8
EC Number
2.7.7.-
Pubmed
19121390
26227816
22118469
26354079
24845553
25830018
+ More
26108605 25315136 26760975 25474469 25348373 21282665 20798317 23537049 28648823 21719571 24438588 25244985 12364791 17994087 15632085 20966253 10731132 11297519 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 30249741 24508170 17550304 22936249 25401762 21292972 20566863 29403074 22293439 23254933 23594743
26108605 25315136 26760975 25474469 25348373 21282665 20798317 23537049 28648823 21719571 24438588 25244985 12364791 17994087 15632085 20966253 10731132 11297519 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 30249741 24508170 17550304 22936249 25401762 21292972 20566863 29403074 22293439 23254933 23594743
EMBL
BABH01002317
ODYU01011852
SOQ57834.1
JTDY01000113
KOB78782.1
NWSH01000359
+ More
PCG77017.1 AGBW02014369 OWR41775.1 KQ459604 KPI92454.1 KK852618 KDR20130.1 GEDC01016245 GEDC01004196 JAS21053.1 JAS33102.1 GBXI01017133 JAC97158.1 JRES01001060 KNC25926.1 FX985855 BBA93742.1 GBBI01003801 JAC14911.1 GAKP01021079 GAKP01021077 JAC37873.1 GDHF01008885 JAI43429.1 GDHF01008483 JAI43831.1 GDHF01015728 GDHF01014261 JAI36586.1 JAI38053.1 GFDL01014570 JAV20475.1 JXJN01001739 DS231969 EDS29710.1 KQ982959 KYQ48769.1 CCAG010019524 KQ976435 KYM87069.1 KQ414646 KOC66420.1 ACPB03004679 ACPB03004680 GL764292 EFZ17938.1 GL435225 EFN73756.1 APGK01052855 KB741216 ENN72480.1 KB632404 ERL95063.1 NNAY01001672 OXU23261.1 GL888181 EGI65810.1 CP012525 ALC43278.1 KQ978615 KYN29814.1 ATLV01022053 KE525327 KFB47136.1 AXCM01004283 GDRN01098677 JAI58906.1 AAAB01008960 EAA11364.4 CH940647 EDW69446.1 CH379070 EAL30303.2 CH963847 EDW73374.1 UFQS01000305 UFQT01000305 SSX02676.1 SSX23050.1 QCYY01000775 ROT82788.1 CH902618 KPU78063.1 EDV39486.1 AE014296 AB049435 AAF47401.1 BAB15801.1 QOIP01000010 RLU17933.1 AXCN02000418 BT031311 ABY21724.1 KK107348 EZA52210.1 LBMM01003755 KMQ93172.1 CH916366 EDV96067.1 CM000159 EDW92739.1 CH933809 EDW18052.2 CH954178 EDV50065.1 CM002912 KMY96701.1 APCN01000100 GBHO01029437 GBHO01029436 JAG14167.1 JAG14168.1 GBRD01009661 GBRD01009659 GBRD01009658 JAG56163.1 OUUW01000012 SPP87497.1 GL732583 EFX74425.1 GDIQ01032457 JAN62280.1 GDIQ01258816 JAJ92908.1 DS235335 EEB14876.1 GDIP01138622 JAL65092.1 GDIQ01224378 JAK27347.1 JH431842 PYGN01000186 PSN51751.1 AKHW03001628 KYO41428.1 KYO41427.1 PZQS01000002 PVD37086.1 AMQN01008979 KB304272 ELU02195.1 BX248325
PCG77017.1 AGBW02014369 OWR41775.1 KQ459604 KPI92454.1 KK852618 KDR20130.1 GEDC01016245 GEDC01004196 JAS21053.1 JAS33102.1 GBXI01017133 JAC97158.1 JRES01001060 KNC25926.1 FX985855 BBA93742.1 GBBI01003801 JAC14911.1 GAKP01021079 GAKP01021077 JAC37873.1 GDHF01008885 JAI43429.1 GDHF01008483 JAI43831.1 GDHF01015728 GDHF01014261 JAI36586.1 JAI38053.1 GFDL01014570 JAV20475.1 JXJN01001739 DS231969 EDS29710.1 KQ982959 KYQ48769.1 CCAG010019524 KQ976435 KYM87069.1 KQ414646 KOC66420.1 ACPB03004679 ACPB03004680 GL764292 EFZ17938.1 GL435225 EFN73756.1 APGK01052855 KB741216 ENN72480.1 KB632404 ERL95063.1 NNAY01001672 OXU23261.1 GL888181 EGI65810.1 CP012525 ALC43278.1 KQ978615 KYN29814.1 ATLV01022053 KE525327 KFB47136.1 AXCM01004283 GDRN01098677 JAI58906.1 AAAB01008960 EAA11364.4 CH940647 EDW69446.1 CH379070 EAL30303.2 CH963847 EDW73374.1 UFQS01000305 UFQT01000305 SSX02676.1 SSX23050.1 QCYY01000775 ROT82788.1 CH902618 KPU78063.1 EDV39486.1 AE014296 AB049435 AAF47401.1 BAB15801.1 QOIP01000010 RLU17933.1 AXCN02000418 BT031311 ABY21724.1 KK107348 EZA52210.1 LBMM01003755 KMQ93172.1 CH916366 EDV96067.1 CM000159 EDW92739.1 CH933809 EDW18052.2 CH954178 EDV50065.1 CM002912 KMY96701.1 APCN01000100 GBHO01029437 GBHO01029436 JAG14167.1 JAG14168.1 GBRD01009661 GBRD01009659 GBRD01009658 JAG56163.1 OUUW01000012 SPP87497.1 GL732583 EFX74425.1 GDIQ01032457 JAN62280.1 GDIQ01258816 JAJ92908.1 DS235335 EEB14876.1 GDIP01138622 JAL65092.1 GDIQ01224378 JAK27347.1 JH431842 PYGN01000186 PSN51751.1 AKHW03001628 KYO41428.1 KYO41427.1 PZQS01000002 PVD37086.1 AMQN01008979 KB304272 ELU02195.1 BX248325
Proteomes
UP000005204
UP000037510
UP000218220
UP000007151
UP000053268
UP000027135
+ More
UP000037069 UP000095301 UP000091820 UP000092445 UP000095300 UP000079169 UP000092443 UP000092460 UP000002320 UP000075809 UP000092444 UP000078200 UP000078540 UP000192221 UP000053825 UP000015103 UP000069272 UP000000311 UP000019118 UP000030742 UP000215335 UP000007755 UP000092553 UP000075880 UP000078492 UP000075885 UP000030765 UP000075883 UP000076408 UP000007062 UP000008792 UP000001819 UP000007798 UP000075882 UP000075920 UP000283509 UP000007801 UP000000803 UP000279307 UP000075886 UP000053097 UP000075903 UP000075881 UP000036403 UP000001070 UP000076407 UP000002282 UP000075884 UP000009192 UP000008711 UP000005203 UP000075840 UP000075902 UP000268350 UP000000305 UP000009046 UP000245037 UP000050525 UP000245119 UP000014760 UP000000437
UP000037069 UP000095301 UP000091820 UP000092445 UP000095300 UP000079169 UP000092443 UP000092460 UP000002320 UP000075809 UP000092444 UP000078200 UP000078540 UP000192221 UP000053825 UP000015103 UP000069272 UP000000311 UP000019118 UP000030742 UP000215335 UP000007755 UP000092553 UP000075880 UP000078492 UP000075885 UP000030765 UP000075883 UP000076408 UP000007062 UP000008792 UP000001819 UP000007798 UP000075882 UP000075920 UP000283509 UP000007801 UP000000803 UP000279307 UP000075886 UP000053097 UP000075903 UP000075881 UP000036403 UP000001070 UP000076407 UP000002282 UP000075884 UP000009192 UP000008711 UP000005203 UP000075840 UP000075902 UP000268350 UP000000305 UP000009046 UP000245037 UP000050525 UP000245119 UP000014760 UP000000437
Pfam
Interpro
IPR017961
DNA_pol_Y-fam_little_finger
+ More
IPR036775 DNA_pol_Y-fam_lit_finger_sf
IPR031991 Rev1_C
IPR012112 REV1
IPR038401 Rev1_C_sf
IPR001126 UmuC
IPR001357 BRCT_dom
IPR024728 PolY_HhH_motif
IPR036420 BRCT_dom_sf
IPR025527 HUWE1/Rev1_UBM
IPR036236 Znf_C2H2_sf
IPR013087 Znf_C2H2_type
IPR018108 Mitochondrial_sb/sol_carrier
IPR023395 Mt_carrier_dom_sf
IPR013717 PIG-P
IPR022880 DNApol_IV
IPR036775 DNA_pol_Y-fam_lit_finger_sf
IPR031991 Rev1_C
IPR012112 REV1
IPR038401 Rev1_C_sf
IPR001126 UmuC
IPR001357 BRCT_dom
IPR024728 PolY_HhH_motif
IPR036420 BRCT_dom_sf
IPR025527 HUWE1/Rev1_UBM
IPR036236 Znf_C2H2_sf
IPR013087 Znf_C2H2_type
IPR018108 Mitochondrial_sb/sol_carrier
IPR023395 Mt_carrier_dom_sf
IPR013717 PIG-P
IPR022880 DNApol_IV
CDD
ProteinModelPortal
H9JFL8
A0A2H1WZM8
A0A0L7LTL5
A0A2A4JYB7
A0A212EJX8
A0A194PIA3
+ More
A0A067R8Z4 A0A1B6D5V3 A0A0A1WDT0 A0A0L0C0Z9 A0A1I8ND87 A0A2Z5TZG2 A0A1A9WC86 A0A1A9ZMM3 A0A1I8PNV4 A0A3Q0JLE4 A0A023F1M3 A0A034V8J5 A0A0K8VX46 A0A0K8VYB8 A0A0K8VCJ4 A0A1Q3EYU1 A0A1A9XPD7 A0A1B0AQ86 B0WKB9 A0A151WLK4 A0A1B0FJR5 A0A1A9V359 A0A195BNY4 A0A1W4W2L6 A0A0L7R6C0 T1HD53 E9INC1 A0A182FCB0 E1ZYK2 N6TT88 U4URC7 A0A232EXZ3 F4WJ40 A0A0M4E8N8 A0A182JB10 A0A151JRB0 A0A182PV68 A0A084WA92 A0A182MNF0 A0A0P4VTD3 A0A182YD29 Q7Q6B2 B4LGK8 Q29DW0 B4MLB6 A0A182KPP2 A0A182WKR2 A0A336KCQ3 A0A3R7MPY2 A0A0P8XTL6 B3M507 Q9W0P2 A0A3L8DCS6 A0A182QM37 A9UNE7 A0A026W820 A0A182VH23 A0A182JNL6 A0A0J7KS11 B4J2S6 A0A182X700 B4PCL6 A0A182NIP1 B4L0A6 B3NJA3 A0A088ASQ5 A0A0J9RLC9 A0A182I1Y4 A0A182UC77 A0A0A9X5V3 A0A0K8SS66 A0A3B0JZI7 E9H1D8 A0A0P6GJG4 A0A0P5F857 E0VNC0 A0A0P5SKK5 A0A0P5HQN3 T1J4C2 A0A2P8Z5H3 A0A151NXA6 A0A151NX22 A0A2T7PUI3 R7U747 A0A1W3JQL8 E9QIK8
A0A067R8Z4 A0A1B6D5V3 A0A0A1WDT0 A0A0L0C0Z9 A0A1I8ND87 A0A2Z5TZG2 A0A1A9WC86 A0A1A9ZMM3 A0A1I8PNV4 A0A3Q0JLE4 A0A023F1M3 A0A034V8J5 A0A0K8VX46 A0A0K8VYB8 A0A0K8VCJ4 A0A1Q3EYU1 A0A1A9XPD7 A0A1B0AQ86 B0WKB9 A0A151WLK4 A0A1B0FJR5 A0A1A9V359 A0A195BNY4 A0A1W4W2L6 A0A0L7R6C0 T1HD53 E9INC1 A0A182FCB0 E1ZYK2 N6TT88 U4URC7 A0A232EXZ3 F4WJ40 A0A0M4E8N8 A0A182JB10 A0A151JRB0 A0A182PV68 A0A084WA92 A0A182MNF0 A0A0P4VTD3 A0A182YD29 Q7Q6B2 B4LGK8 Q29DW0 B4MLB6 A0A182KPP2 A0A182WKR2 A0A336KCQ3 A0A3R7MPY2 A0A0P8XTL6 B3M507 Q9W0P2 A0A3L8DCS6 A0A182QM37 A9UNE7 A0A026W820 A0A182VH23 A0A182JNL6 A0A0J7KS11 B4J2S6 A0A182X700 B4PCL6 A0A182NIP1 B4L0A6 B3NJA3 A0A088ASQ5 A0A0J9RLC9 A0A182I1Y4 A0A182UC77 A0A0A9X5V3 A0A0K8SS66 A0A3B0JZI7 E9H1D8 A0A0P6GJG4 A0A0P5F857 E0VNC0 A0A0P5SKK5 A0A0P5HQN3 T1J4C2 A0A2P8Z5H3 A0A151NXA6 A0A151NX22 A0A2T7PUI3 R7U747 A0A1W3JQL8 E9QIK8
PDB
3GQC
E-value=4.29673e-112,
Score=1038
Ontologies
GO
Topology
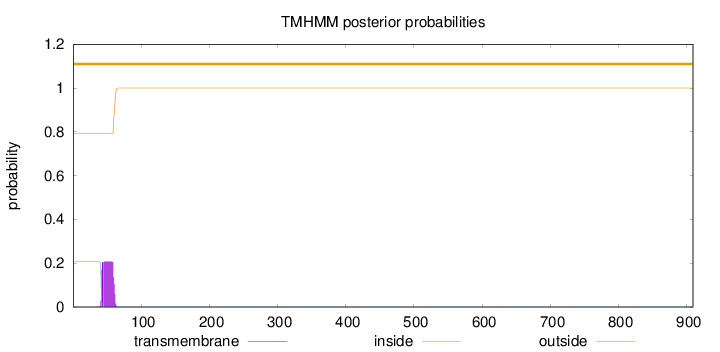
Subcellular location
Nucleus
Length:
909
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
4.04232999999999
Exp number, first 60 AAs:
3.85011
Total prob of N-in:
0.20731
outside
1 - 909
Population Genetic Test Statistics
Pi
212.873167
Theta
240.322627
Tajima's D
-1.010608
CLR
2964.282336
CSRT
0.137943102844858
Interpretation
Uncertain
