Gene
KWMTBOMO11057
Pre Gene Modal
BGIBMGA008544
Annotation
PREDICTED:_tubulin-specific_chaperone_cofactor_E-like_protein_isoform_X2_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 1.635 Nuclear Reliability : 1.761
Sequence
CDS
ATGCCGTCGCTTTTGGAAGCCCTTGAGCGCAAATACGGCGCTAAGGGCGAAGTAAATCCATCAATTGACGACATGCCAGTTGCCATATTTGTGCCTAAACGCTCCCCGAGGCTTAGCGTACCCACACTGCTAGTACTCAACGATTGCGATATAGACTGTGCCGGAGATGCATGTATACTAGCAGATAAATGTGCGGACGTTGTTGAACTTGATCTAGCTAACAATAAATTAACTGAATGGCAAGAGGTGTTTGCAATATTAGAGCAGACTCCACGAGTTCGATTCCTTAATTTGAGTTTTAACAGGCTTTCGGCTCAGATTCAAGCAGCCCAGTCATTACGGCCCAGATTGGATAGTTTGAGCTATCTGGTCCTTAACTCTACCTATGTTAGTTGGCCAAGTGTTCATTCGTTACTCAAGGCTCTCCCAGCTCTCGAAGAACTGCACTTGAGCCTCAATGAGTATTCGTATGTGAATTTAACTGGACAAGAAGTCGACGAAAAGGATCGTTGCAATGCATGTTCCCGTTTGAACGTTGCTGATGTCACTGAGCCAGTTCATGAACAGCTAAAGAAACTTCATTTCTCCGGGAATCCAGTCAGCAGCTGGTGGGAGATTTCCAAATTGGGCTATGCCTTCCCCAATCTGGAAACGCTCCTCGTTAATGAATGTCATTTAACAACCTTAGAGCCGGATCCTTGCGAGAAGTGTGGAGACGAAAACGGGAACAAAAAGAGTCACGATGCTTTCTCACATTTACGCTTCCTTAATCTGAACAACACCGGTTTGGCAACATGGGATGAAGTGGATCGGTTGGCGCGGTTTCCTGCCCTGAAAAGTTTACGAGTTCAGGGTTGGCCGCTGTGGGAACGCGTCGAGTCAACAGAGCACGAGAGACGATTGCTTTTAGTGGCTCGTCTCCCTCACATAAGGACCTTGAACGGAGGAGGGGCGGTCCCTGTTGAAGAACGAGATGCTGCAGAGAGGGCCTTCATCAGATATTACATGGAAAAGCCAGAGGCCGATCGACCGGATAGATACTGGGAGCTGGTGCACGTGCACGGCAAGCTCGATCCGCTGGTGTCCGTGGACCTGCGGCCCGAGAAGCGCGTGCAGATCACGTTCACGTGCGGGGACACGAGCGAAGTGCGCACCGTCGACGTTTACAGGACTGTTTCTGATCTAAAAACAAGATTGGAGCGGTTAGCTGGCTTCCCGGCTGCGAAGATGCGGCTCTTTTACGTGGACCAAGAACTGAGAGATACGCAGGGTCCCGAAGAGATGAAGTACCCTACGAAGCAGCTATACTCGTACAACATCAGGTCGGGGGATGAGATCATCATCGACTCGAAACTGAAGCACTCTATATCGGCCAACTCGACCGCCTGA
Protein
MPSLLEALERKYGAKGEVNPSIDDMPVAIFVPKRSPRLSVPTLLVLNDCDIDCAGDACILADKCADVVELDLANNKLTEWQEVFAILEQTPRVRFLNLSFNRLSAQIQAAQSLRPRLDSLSYLVLNSTYVSWPSVHSLLKALPALEELHLSLNEYSYVNLTGQEVDEKDRCNACSRLNVADVTEPVHEQLKKLHFSGNPVSSWWEISKLGYAFPNLETLLVNECHLTTLEPDPCEKCGDENGNKKSHDAFSHLRFLNLNNTGLATWDEVDRLARFPALKSLRVQGWPLWERVESTEHERRLLLVARLPHIRTLNGGGAVPVEERDAAERAFIRYYMEKPEADRPDRYWELVHVHGKLDPLVSVDLRPEKRVQITFTCGDTSEVRTVDVYRTVSDLKTRLERLAGFPAAKMRLFYVDQELRDTQGPEEMKYPTKQLYSYNIRSGDEIIIDSKLKHSISANSTA
Summary
Similarity
Belongs to the potassium channel family.
Uniprot
A0A1E1WPM1
A0A2H1WXN7
A0A2A4JYA8
A0A1E1WU68
A0A0N0PBT0
S4PXG4
+ More
A0A212FAJ6 A0A3S2M5B6 A0A194PMV5 A0A0L7LU89 D2A1Y4 U4UKC2 J3JWV6 A0A1L8E5X2 A0A1B0CX35 U5EXJ5 A0A1Y1MS15 B0X188 A0A023ET00 A0A1Q3FKD9 Q17CU4 A0A336MA02 A0A336MHJ0 A0A151X747 A0A2J7PGD2 A0A182GHI7 A0A026WKS0 A0A195ES82 E2BHG6 A0A1B6DSK6 A0A154P4T6 A0A0M8ZWL0 A0A151JQU4 A0A151I364 A0A158NSD2 A0A3L8DAC9 F4W5L4 E2AEE5 A0A087ZWA8 A0A0L7QV09 A0A195D571 E9IYF1 A0A067QWB7 A0A1J1IFV5 A0A1A9W2G9 K7J345 A0A2H8TP41 J9K6N8 A0A1A9ZGX1 A0A2S2QU29 Q7K549 A0A1A9VD05 W5JLY1 C6SUV9 A0A2M4BL03 E0VIM4 B4QIA0 B4HME2 B3MBF3 B3N6H4 A0A1I8PBM8 Q7QHD6 B4NX61 A0A1W4VCP0 Q290I9 A0A182LYF2 B4KN73 A0A182J9Z7 A0A182RW55 A0A2M3Z0M0 A0A2M3Z0P6 A0A1A9XAB4 B4GBS7 A0A182FW50 T1PBR7 A0A1I8NE40 A0A3B0JSJ4 B4MIM1 A0A182Q1F6 A0A0A9X3S2 B4LKB5 A0A182N2A2 A0A0M5IX47 B4J6V6 A0A0N7Z958 T1I0V1 A0A023F2S4 A0A1B6GSG0 A0A1B0FNK7 A0A069DZ16 A0A2M4A475 A0A1B6LPC6 A0A182KL14 A0A2R7WGU9 A0A0C9Q0W7 A0A1Y1MWW4 A0A2M4A693 A0A182WDK0
A0A212FAJ6 A0A3S2M5B6 A0A194PMV5 A0A0L7LU89 D2A1Y4 U4UKC2 J3JWV6 A0A1L8E5X2 A0A1B0CX35 U5EXJ5 A0A1Y1MS15 B0X188 A0A023ET00 A0A1Q3FKD9 Q17CU4 A0A336MA02 A0A336MHJ0 A0A151X747 A0A2J7PGD2 A0A182GHI7 A0A026WKS0 A0A195ES82 E2BHG6 A0A1B6DSK6 A0A154P4T6 A0A0M8ZWL0 A0A151JQU4 A0A151I364 A0A158NSD2 A0A3L8DAC9 F4W5L4 E2AEE5 A0A087ZWA8 A0A0L7QV09 A0A195D571 E9IYF1 A0A067QWB7 A0A1J1IFV5 A0A1A9W2G9 K7J345 A0A2H8TP41 J9K6N8 A0A1A9ZGX1 A0A2S2QU29 Q7K549 A0A1A9VD05 W5JLY1 C6SUV9 A0A2M4BL03 E0VIM4 B4QIA0 B4HME2 B3MBF3 B3N6H4 A0A1I8PBM8 Q7QHD6 B4NX61 A0A1W4VCP0 Q290I9 A0A182LYF2 B4KN73 A0A182J9Z7 A0A182RW55 A0A2M3Z0M0 A0A2M3Z0P6 A0A1A9XAB4 B4GBS7 A0A182FW50 T1PBR7 A0A1I8NE40 A0A3B0JSJ4 B4MIM1 A0A182Q1F6 A0A0A9X3S2 B4LKB5 A0A182N2A2 A0A0M5IX47 B4J6V6 A0A0N7Z958 T1I0V1 A0A023F2S4 A0A1B6GSG0 A0A1B0FNK7 A0A069DZ16 A0A2M4A475 A0A1B6LPC6 A0A182KL14 A0A2R7WGU9 A0A0C9Q0W7 A0A1Y1MWW4 A0A2M4A693 A0A182WDK0
Pubmed
26354079
23622113
22118469
26227816
18362917
19820115
+ More
23537049 22516182 28004739 24945155 17510324 26483478 24508170 20798317 21347285 30249741 21719571 21282665 24845553 20075255 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20920257 23761445 20566863 17994087 22936249 12364791 17550304 15632085 25315136 25401762 26823975 27129103 25474469 26334808 20966253
23537049 22516182 28004739 24945155 17510324 26483478 24508170 20798317 21347285 30249741 21719571 21282665 24845553 20075255 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20920257 23761445 20566863 17994087 22936249 12364791 17550304 15632085 25315136 25401762 26823975 27129103 25474469 26334808 20966253
EMBL
GDQN01010136
GDQN01002095
JAT80918.1
JAT88959.1
ODYU01011852
SOQ57835.1
+ More
NWSH01000359 PCG77005.1 GDQN01000540 GDQN01000233 JAT90514.1 JAT90821.1 KQ460813 KPJ12152.1 GAIX01005083 JAA87477.1 AGBW02009455 OWR50764.1 RSAL01000035 RVE51324.1 KQ459604 KPI92455.1 JTDY01000113 KOB78786.1 KQ971338 EFA02067.1 KB632390 ERL94529.1 BT127724 AEE62686.1 GFDF01000008 JAV14076.1 AJWK01033180 GANO01000057 JAB59814.1 GEZM01024211 JAV87868.1 DS232256 EDS38537.1 GAPW01001176 JAC12422.1 GFDL01007073 JAV27972.1 CH477304 EAT44178.1 UFQS01000769 UFQT01000769 SSX06740.1 SSX27085.1 UFQT01001095 SSX28891.1 KQ982459 KYQ56149.1 NEVH01025635 PNF15389.1 JXUM01011798 KQ560338 KXJ82969.1 KK107167 EZA56266.1 KQ981993 KYN31031.1 GL448287 EFN84935.1 GEDC01030871 GEDC01028021 GEDC01020684 GEDC01017087 GEDC01008631 GEDC01000902 JAS06427.1 JAS09277.1 JAS16614.1 JAS20211.1 JAS28667.1 JAS36396.1 KQ434819 KZC06959.1 KQ435821 KOX72407.1 KQ978642 KYN29490.1 KQ976543 KYM80972.1 ADTU01024828 QOIP01000010 RLU17455.1 GL887655 EGI70511.1 GL438828 EFN68197.1 KQ414727 KOC62497.1 KQ976881 KYN07574.1 GL767018 EFZ14354.1 KK853289 KDR08895.1 CVRI01000048 CRK99133.1 GFXV01004138 MBW15943.1 ABLF02016657 ABLF02016659 ABLF02016660 ABLF02055163 GGMS01012048 MBY81251.1 AE013599 AY051458 AAF58828.1 AAK92882.1 AAM68775.1 AGB93386.1 ADMH02001151 ETN63895.1 BT088951 ACT79357.1 GGFJ01004490 MBW53631.1 DS235201 EEB13230.1 CM000362 CM002911 EDX06460.1 KMY92723.1 CH480816 EDW47219.1 CH902619 EDV36078.1 CH954177 EDV58143.1 AAAB01008816 EAA05315.4 CM000157 EDW89622.1 CM000071 EAL25373.1 AXCM01001629 CH933808 EDW08900.1 GGFM01001328 MBW22079.1 GGFM01001342 MBW22093.1 CH479181 EDW32335.1 KA645555 AFP60184.1 OUUW01000001 SPP74078.1 CH963719 EDW71960.1 AXCN02001730 GBHO01028225 GBHO01028222 GBRD01017094 GBRD01017093 GDHC01021823 GDHC01017359 JAG15379.1 JAG15382.1 JAG48733.1 JAP96805.1 JAQ01270.1 CH940648 EDW61706.1 CP012524 ALC40581.1 CH916367 EDW02037.1 GDKW01001467 JAI55128.1 ACPB03017977 GBBI01003082 JAC15630.1 GECZ01004410 JAS65359.1 CCAG010001879 GBGD01001425 JAC87464.1 GGFK01002209 MBW35530.1 GEBQ01014375 JAT25602.1 KK854788 PTY18739.1 GBYB01007553 JAG77320.1 GEZM01024212 JAV87867.1 GGFK01002827 MBW36148.1
NWSH01000359 PCG77005.1 GDQN01000540 GDQN01000233 JAT90514.1 JAT90821.1 KQ460813 KPJ12152.1 GAIX01005083 JAA87477.1 AGBW02009455 OWR50764.1 RSAL01000035 RVE51324.1 KQ459604 KPI92455.1 JTDY01000113 KOB78786.1 KQ971338 EFA02067.1 KB632390 ERL94529.1 BT127724 AEE62686.1 GFDF01000008 JAV14076.1 AJWK01033180 GANO01000057 JAB59814.1 GEZM01024211 JAV87868.1 DS232256 EDS38537.1 GAPW01001176 JAC12422.1 GFDL01007073 JAV27972.1 CH477304 EAT44178.1 UFQS01000769 UFQT01000769 SSX06740.1 SSX27085.1 UFQT01001095 SSX28891.1 KQ982459 KYQ56149.1 NEVH01025635 PNF15389.1 JXUM01011798 KQ560338 KXJ82969.1 KK107167 EZA56266.1 KQ981993 KYN31031.1 GL448287 EFN84935.1 GEDC01030871 GEDC01028021 GEDC01020684 GEDC01017087 GEDC01008631 GEDC01000902 JAS06427.1 JAS09277.1 JAS16614.1 JAS20211.1 JAS28667.1 JAS36396.1 KQ434819 KZC06959.1 KQ435821 KOX72407.1 KQ978642 KYN29490.1 KQ976543 KYM80972.1 ADTU01024828 QOIP01000010 RLU17455.1 GL887655 EGI70511.1 GL438828 EFN68197.1 KQ414727 KOC62497.1 KQ976881 KYN07574.1 GL767018 EFZ14354.1 KK853289 KDR08895.1 CVRI01000048 CRK99133.1 GFXV01004138 MBW15943.1 ABLF02016657 ABLF02016659 ABLF02016660 ABLF02055163 GGMS01012048 MBY81251.1 AE013599 AY051458 AAF58828.1 AAK92882.1 AAM68775.1 AGB93386.1 ADMH02001151 ETN63895.1 BT088951 ACT79357.1 GGFJ01004490 MBW53631.1 DS235201 EEB13230.1 CM000362 CM002911 EDX06460.1 KMY92723.1 CH480816 EDW47219.1 CH902619 EDV36078.1 CH954177 EDV58143.1 AAAB01008816 EAA05315.4 CM000157 EDW89622.1 CM000071 EAL25373.1 AXCM01001629 CH933808 EDW08900.1 GGFM01001328 MBW22079.1 GGFM01001342 MBW22093.1 CH479181 EDW32335.1 KA645555 AFP60184.1 OUUW01000001 SPP74078.1 CH963719 EDW71960.1 AXCN02001730 GBHO01028225 GBHO01028222 GBRD01017094 GBRD01017093 GDHC01021823 GDHC01017359 JAG15379.1 JAG15382.1 JAG48733.1 JAP96805.1 JAQ01270.1 CH940648 EDW61706.1 CP012524 ALC40581.1 CH916367 EDW02037.1 GDKW01001467 JAI55128.1 ACPB03017977 GBBI01003082 JAC15630.1 GECZ01004410 JAS65359.1 CCAG010001879 GBGD01001425 JAC87464.1 GGFK01002209 MBW35530.1 GEBQ01014375 JAT25602.1 KK854788 PTY18739.1 GBYB01007553 JAG77320.1 GEZM01024212 JAV87867.1 GGFK01002827 MBW36148.1
Proteomes
UP000218220
UP000053240
UP000007151
UP000283053
UP000053268
UP000037510
+ More
UP000007266 UP000030742 UP000092461 UP000002320 UP000008820 UP000075809 UP000235965 UP000069940 UP000249989 UP000053097 UP000078541 UP000008237 UP000076502 UP000053105 UP000078492 UP000078540 UP000005205 UP000279307 UP000007755 UP000000311 UP000005203 UP000053825 UP000078542 UP000027135 UP000183832 UP000091820 UP000002358 UP000007819 UP000092445 UP000000803 UP000078200 UP000000673 UP000009046 UP000000304 UP000001292 UP000007801 UP000008711 UP000095300 UP000007062 UP000002282 UP000192221 UP000001819 UP000075883 UP000009192 UP000075880 UP000075900 UP000092443 UP000008744 UP000069272 UP000095301 UP000268350 UP000007798 UP000075886 UP000008792 UP000075884 UP000092553 UP000001070 UP000015103 UP000092444 UP000075882 UP000075920
UP000007266 UP000030742 UP000092461 UP000002320 UP000008820 UP000075809 UP000235965 UP000069940 UP000249989 UP000053097 UP000078541 UP000008237 UP000076502 UP000053105 UP000078492 UP000078540 UP000005205 UP000279307 UP000007755 UP000000311 UP000005203 UP000053825 UP000078542 UP000027135 UP000183832 UP000091820 UP000002358 UP000007819 UP000092445 UP000000803 UP000078200 UP000000673 UP000009046 UP000000304 UP000001292 UP000007801 UP000008711 UP000095300 UP000007062 UP000002282 UP000192221 UP000001819 UP000075883 UP000009192 UP000075880 UP000075900 UP000092443 UP000008744 UP000069272 UP000095301 UP000268350 UP000007798 UP000075886 UP000008792 UP000075884 UP000092553 UP000001070 UP000015103 UP000092444 UP000075882 UP000075920
Interpro
SUPFAM
SSF54236
SSF54236
Gene 3D
ProteinModelPortal
A0A1E1WPM1
A0A2H1WXN7
A0A2A4JYA8
A0A1E1WU68
A0A0N0PBT0
S4PXG4
+ More
A0A212FAJ6 A0A3S2M5B6 A0A194PMV5 A0A0L7LU89 D2A1Y4 U4UKC2 J3JWV6 A0A1L8E5X2 A0A1B0CX35 U5EXJ5 A0A1Y1MS15 B0X188 A0A023ET00 A0A1Q3FKD9 Q17CU4 A0A336MA02 A0A336MHJ0 A0A151X747 A0A2J7PGD2 A0A182GHI7 A0A026WKS0 A0A195ES82 E2BHG6 A0A1B6DSK6 A0A154P4T6 A0A0M8ZWL0 A0A151JQU4 A0A151I364 A0A158NSD2 A0A3L8DAC9 F4W5L4 E2AEE5 A0A087ZWA8 A0A0L7QV09 A0A195D571 E9IYF1 A0A067QWB7 A0A1J1IFV5 A0A1A9W2G9 K7J345 A0A2H8TP41 J9K6N8 A0A1A9ZGX1 A0A2S2QU29 Q7K549 A0A1A9VD05 W5JLY1 C6SUV9 A0A2M4BL03 E0VIM4 B4QIA0 B4HME2 B3MBF3 B3N6H4 A0A1I8PBM8 Q7QHD6 B4NX61 A0A1W4VCP0 Q290I9 A0A182LYF2 B4KN73 A0A182J9Z7 A0A182RW55 A0A2M3Z0M0 A0A2M3Z0P6 A0A1A9XAB4 B4GBS7 A0A182FW50 T1PBR7 A0A1I8NE40 A0A3B0JSJ4 B4MIM1 A0A182Q1F6 A0A0A9X3S2 B4LKB5 A0A182N2A2 A0A0M5IX47 B4J6V6 A0A0N7Z958 T1I0V1 A0A023F2S4 A0A1B6GSG0 A0A1B0FNK7 A0A069DZ16 A0A2M4A475 A0A1B6LPC6 A0A182KL14 A0A2R7WGU9 A0A0C9Q0W7 A0A1Y1MWW4 A0A2M4A693 A0A182WDK0
A0A212FAJ6 A0A3S2M5B6 A0A194PMV5 A0A0L7LU89 D2A1Y4 U4UKC2 J3JWV6 A0A1L8E5X2 A0A1B0CX35 U5EXJ5 A0A1Y1MS15 B0X188 A0A023ET00 A0A1Q3FKD9 Q17CU4 A0A336MA02 A0A336MHJ0 A0A151X747 A0A2J7PGD2 A0A182GHI7 A0A026WKS0 A0A195ES82 E2BHG6 A0A1B6DSK6 A0A154P4T6 A0A0M8ZWL0 A0A151JQU4 A0A151I364 A0A158NSD2 A0A3L8DAC9 F4W5L4 E2AEE5 A0A087ZWA8 A0A0L7QV09 A0A195D571 E9IYF1 A0A067QWB7 A0A1J1IFV5 A0A1A9W2G9 K7J345 A0A2H8TP41 J9K6N8 A0A1A9ZGX1 A0A2S2QU29 Q7K549 A0A1A9VD05 W5JLY1 C6SUV9 A0A2M4BL03 E0VIM4 B4QIA0 B4HME2 B3MBF3 B3N6H4 A0A1I8PBM8 Q7QHD6 B4NX61 A0A1W4VCP0 Q290I9 A0A182LYF2 B4KN73 A0A182J9Z7 A0A182RW55 A0A2M3Z0M0 A0A2M3Z0P6 A0A1A9XAB4 B4GBS7 A0A182FW50 T1PBR7 A0A1I8NE40 A0A3B0JSJ4 B4MIM1 A0A182Q1F6 A0A0A9X3S2 B4LKB5 A0A182N2A2 A0A0M5IX47 B4J6V6 A0A0N7Z958 T1I0V1 A0A023F2S4 A0A1B6GSG0 A0A1B0FNK7 A0A069DZ16 A0A2M4A475 A0A1B6LPC6 A0A182KL14 A0A2R7WGU9 A0A0C9Q0W7 A0A1Y1MWW4 A0A2M4A693 A0A182WDK0
PDB
5YXM
E-value=7.91576e-05,
Score=110
Ontologies
GO
PANTHER
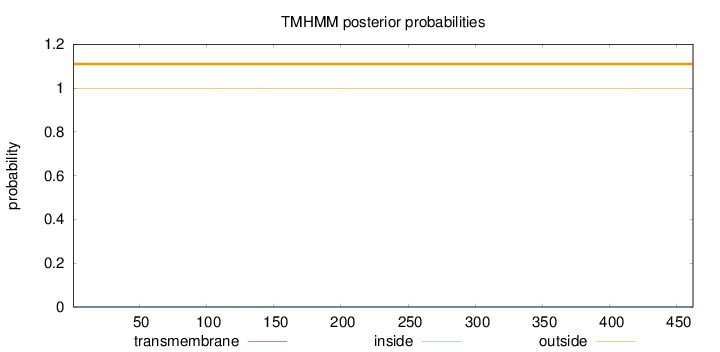
Topology
Length:
462
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00137
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00064
outside
1 - 462
Population Genetic Test Statistics
Pi
267.278497
Theta
174.958986
Tajima's D
0.997956
CLR
0.32114
CSRT
0.659917004149792
Interpretation
Uncertain
