Gene
KWMTBOMO11056 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008316
Annotation
PREDICTED:_persulfide_dioxygenase_ETHE1?_mitochondrial_[Papilio_polytes]
Location in the cell
Mitochondrial Reliability : 2.209
Sequence
CDS
ATGTTGCAATCATTGGTGTTTGGTGGACGGAAGGCGGCTGATAGATACAAACTTTCAAATTTATTATTTCATGTCAGAACTATGTCAGCAGTTCATAAAGACACCACTTTCTTTTTCAGACAGTTATTTGATACAGTGAGTAGCACATATACATATTTACTTGGAGATAAGAAGACTGGTGAAGTGGTCCTCATAGATCCAGTTTTAGAACACGCCGAGAGGGATGCTAAGTTAATACAGGAACTTGGCTTTAAACTCATTTATGCTATGAACACACACATGCACGCCGACCATATAACCGGCACTGGGAAGTTGAAGTCACTCGTACCGGGTACTAAAAGCGTCATAGGCAAGGATTCGGGTGCCCAAGCCGATATCCACCTGCTCGACGGTGACCTGGTGCGGTTCGGGAGCCACGAACTCCTCGCTGCTGCGACTCCGGGCCACACTAATGGATGCTTGACTTACATATGTCACGAACAGTCATTGGCGTTCACTGGTGACACCCTCTTGATCCGGGGTTGCGGCCGCACCGATTTCCAGGAGGGTAACTCTGAGACCCTCTACAAATCGGTCCACAATAGAATATTCACGTTGCCCGACGAGTACGTGCTGTATCCCGCGCACGATTACCGAGGGCAGACAGCGACATCTGTCGCCGAAGAGAAGAAATACAATCCCAGATTGACGAAGTCGCTTGCCGAGTTCGTTGAGATCATGGATAATTTGAATTTGCCGTATCCGAAAATGATAAATAAAGCGGTGCCGGCGAACCGTGTTTGCGGATTATTTTGA
Protein
MLQSLVFGGRKAADRYKLSNLLFHVRTMSAVHKDTTFFFRQLFDTVSSTYTYLLGDKKTGEVVLIDPVLEHAERDAKLIQELGFKLIYAMNTHMHADHITGTGKLKSLVPGTKSVIGKDSGAQADIHLLDGDLVRFGSHELLAAATPGHTNGCLTYICHEQSLAFTGDTLLIRGCGRTDFQEGNSETLYKSVHNRIFTLPDEYVLYPAHDYRGQTATSVAEEKKYNPRLTKSLAEFVEIMDNLNLPYPKMINKAVPANRVCGLF
Summary
Uniprot
H9JFL9
A0A194PGE0
A0A2A4JYA9
A0A3S2LPT8
A0A2H1WFB4
A0A023EMQ1
+ More
Q16V29 Q1HR84 A0A182XKE3 A0A182JJL5 A0A182VXS1 A0A182HJK1 Q7PPW0 A0A1Q3FYK0 A0A182VMD3 A0A2M3ZKV2 A0A1S4FN35 A0A182QI82 W5JLL9 A0A182RQQ5 A0A182MN27 A0A2M3ZBV3 A0A2M4BUI4 A0A182KWQ8 A0A2M4BUN5 A0A182FP83 A0A084VC25 A0A2M4ALW4 A0A182P8P6 A0A2M4ALV5 A0A2M4ALB8 K7J5S8 A0A336MVK9 A0A182NFU4 A0A067QTI4 A0A336MQI3 E0W3Y8 D3TQQ7 A0A182XXA9 A0A1A9UIZ4 A0A182K9S8 A0A1J1HFT1 A0A1I8PJE4 B0XDM9 A0A1B6FJV1 T1PER7 A0A1B6HTX8 A0A1I8NHM9 E2C1H5 A0A1Y1N6W1 A0A2M4AN96 A0A0N7ZC78 A0A158NIL8 A0A2J7PGF2 A0A1A9WP43 A0A1L8E4Q3 A0A0P5WQ95 R4UMF5 A0A0P5HIF7 N6WCA6 A0A3B0J3J3 A0A0P6BS76 A0A336M0A0 A0A023GE00 G3MHK2 Q6DJA2 A0A0P5QCN0 A0A0P5HGY7 A0A0P5J5A2 W8BYG1 A0A1L8E5A6 A0A1B6DAS7 A0A0A1WKG7 A0A0P8XM15 A0A0P6IJ81 A0A182U7B4 E2A6I9 A0A3L8D5C5 A0A0Q9XC68 A0A034WBL6 Q28BZ5 R4WDW4 A0A0L0CND8 A0A0Q9W4E1 A0A154NZN0 A0A1E1X8A1 E9GY10 A0A1B6D9M4 Q8AVQ9 A0A3R7QRL1 A0A3B0IZL4 B5E059 A0A2A3E3R6 A0A088AAM8 A0A0R1DWE9 D2A1Y2 A0A1A9YPH6 Q86PD3 A0A2P2HZ67
Q16V29 Q1HR84 A0A182XKE3 A0A182JJL5 A0A182VXS1 A0A182HJK1 Q7PPW0 A0A1Q3FYK0 A0A182VMD3 A0A2M3ZKV2 A0A1S4FN35 A0A182QI82 W5JLL9 A0A182RQQ5 A0A182MN27 A0A2M3ZBV3 A0A2M4BUI4 A0A182KWQ8 A0A2M4BUN5 A0A182FP83 A0A084VC25 A0A2M4ALW4 A0A182P8P6 A0A2M4ALV5 A0A2M4ALB8 K7J5S8 A0A336MVK9 A0A182NFU4 A0A067QTI4 A0A336MQI3 E0W3Y8 D3TQQ7 A0A182XXA9 A0A1A9UIZ4 A0A182K9S8 A0A1J1HFT1 A0A1I8PJE4 B0XDM9 A0A1B6FJV1 T1PER7 A0A1B6HTX8 A0A1I8NHM9 E2C1H5 A0A1Y1N6W1 A0A2M4AN96 A0A0N7ZC78 A0A158NIL8 A0A2J7PGF2 A0A1A9WP43 A0A1L8E4Q3 A0A0P5WQ95 R4UMF5 A0A0P5HIF7 N6WCA6 A0A3B0J3J3 A0A0P6BS76 A0A336M0A0 A0A023GE00 G3MHK2 Q6DJA2 A0A0P5QCN0 A0A0P5HGY7 A0A0P5J5A2 W8BYG1 A0A1L8E5A6 A0A1B6DAS7 A0A0A1WKG7 A0A0P8XM15 A0A0P6IJ81 A0A182U7B4 E2A6I9 A0A3L8D5C5 A0A0Q9XC68 A0A034WBL6 Q28BZ5 R4WDW4 A0A0L0CND8 A0A0Q9W4E1 A0A154NZN0 A0A1E1X8A1 E9GY10 A0A1B6D9M4 Q8AVQ9 A0A3R7QRL1 A0A3B0IZL4 B5E059 A0A2A3E3R6 A0A088AAM8 A0A0R1DWE9 D2A1Y2 A0A1A9YPH6 Q86PD3 A0A2P2HZ67
Pubmed
19121390
26354079
24945155
26483478
17510324
17204158
+ More
12364791 14747013 17210077 20920257 23761445 20966253 24438588 20075255 24845553 20566863 20353571 25244985 25315136 20798317 28004739 21347285 15632085 17994087 22216098 20431018 24495485 25830018 30249741 18057021 25348373 23691247 26108605 28503490 21292972 27762356 23185243 17550304 18362917 19820115 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
12364791 14747013 17210077 20920257 23761445 20966253 24438588 20075255 24845553 20566863 20353571 25244985 25315136 20798317 28004739 21347285 15632085 17994087 22216098 20431018 24495485 25830018 30249741 18057021 25348373 23691247 26108605 28503490 21292972 27762356 23185243 17550304 18362917 19820115 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
EMBL
BABH01002316
KQ459604
KPI92456.1
NWSH01000359
PCG77007.1
RSAL01000035
+ More
RVE51325.1 ODYU01008303 SOQ51775.1 JXUM01042665 GAPW01002965 KQ561333 JAC10633.1 KXJ78937.1 CH477605 EAT38400.1 EAT38401.1 DQ440210 ABF18243.1 APCN01002178 AAAB01008904 EAA09645.4 GFDL01002437 JAV32608.1 GGFM01008405 MBW29156.1 AXCN02000248 ADMH02000799 ETN65016.1 AXCM01001599 GGFM01005199 MBW25950.1 GGFJ01007584 MBW56725.1 GGFJ01007583 MBW56724.1 ATLV01009983 KE524560 KFB35519.1 GGFK01008465 MBW41786.1 GGFK01008267 MBW41588.1 GGFK01008233 MBW41554.1 AAZX01008684 UFQS01003149 UFQT01003149 SSX15269.1 SSX34644.1 KK853289 KDR08896.1 UFQS01001567 UFQT01001567 SSX11476.1 SSX31043.1 DS235885 EEB20344.1 EZ423759 ADD20035.1 CVRI01000002 CRK86831.1 DS232769 EDS45537.1 GECZ01019310 GECZ01009027 JAS50459.1 JAS60742.1 KA647231 AFP61860.1 GECU01029586 JAS78120.1 GL451937 EFN78151.1 GEZM01011117 JAV93644.1 GGFK01008933 MBW42254.1 GDRN01071433 GDRN01071432 JAI63688.1 ADTU01001813 ADTU01001814 ADTU01001815 NEVH01025635 PNF15416.1 GFDF01000373 JAV13711.1 GDIP01095762 JAM07953.1 KC632476 AGM32290.1 GDIQ01227016 JAK24709.1 CM000071 ENO01784.1 OUUW01000001 SPP73853.1 GDIP01025563 JAM78152.1 UFQS01000280 UFQT01000280 SSX02346.1 SSX22721.1 GBBM01004215 JAC31203.1 JO841353 AEO32970.1 AAMC01089184 AAMC01089185 BC075280 AAH75280.1 GDIQ01119055 JAL32671.1 GDIQ01227017 JAK24708.1 GDIQ01206372 JAK45353.1 GAMC01008220 GAMC01008219 JAB98336.1 GFDF01000372 JAV13712.1 GEDC01014505 JAS22793.1 GBXI01015116 GBXI01003618 JAC99175.1 JAD10674.1 CH902619 KPU75734.1 GDIQ01003590 JAN91147.1 GL437132 EFN70946.1 QOIP01000013 RLU15351.1 CH933808 KRG05209.1 KRG05210.1 KRG05211.1 GAKP01007275 GAKP01007274 GAKP01007273 JAC51677.1 CR942545 CAJ81741.1 AK417914 BAN21129.1 JRES01000160 KNC33732.1 CH940648 KRF79928.1 KRF79929.1 KRF79930.1 KRF79931.1 KQ434786 KZC05125.1 GFAC01003696 JAT95492.1 GL732574 EFX75643.1 GEDC01019086 GEDC01014910 JAS18212.1 JAS22388.1 BC041511 CM004478 AAH41511.1 OCT73259.1 QCYY01001749 ROT75645.1 SPP73854.1 EDY69293.2 KRT02805.1 KZ288392 PBC26340.1 CM000158 KRJ99521.1 KQ971338 EFA02967.1 AE013599 BT003193 BT012447 BT033012 AAF58646.3 AAO24948.1 AAS93718.1 ACD99576.1 IACF01001360 LAB67067.1
RVE51325.1 ODYU01008303 SOQ51775.1 JXUM01042665 GAPW01002965 KQ561333 JAC10633.1 KXJ78937.1 CH477605 EAT38400.1 EAT38401.1 DQ440210 ABF18243.1 APCN01002178 AAAB01008904 EAA09645.4 GFDL01002437 JAV32608.1 GGFM01008405 MBW29156.1 AXCN02000248 ADMH02000799 ETN65016.1 AXCM01001599 GGFM01005199 MBW25950.1 GGFJ01007584 MBW56725.1 GGFJ01007583 MBW56724.1 ATLV01009983 KE524560 KFB35519.1 GGFK01008465 MBW41786.1 GGFK01008267 MBW41588.1 GGFK01008233 MBW41554.1 AAZX01008684 UFQS01003149 UFQT01003149 SSX15269.1 SSX34644.1 KK853289 KDR08896.1 UFQS01001567 UFQT01001567 SSX11476.1 SSX31043.1 DS235885 EEB20344.1 EZ423759 ADD20035.1 CVRI01000002 CRK86831.1 DS232769 EDS45537.1 GECZ01019310 GECZ01009027 JAS50459.1 JAS60742.1 KA647231 AFP61860.1 GECU01029586 JAS78120.1 GL451937 EFN78151.1 GEZM01011117 JAV93644.1 GGFK01008933 MBW42254.1 GDRN01071433 GDRN01071432 JAI63688.1 ADTU01001813 ADTU01001814 ADTU01001815 NEVH01025635 PNF15416.1 GFDF01000373 JAV13711.1 GDIP01095762 JAM07953.1 KC632476 AGM32290.1 GDIQ01227016 JAK24709.1 CM000071 ENO01784.1 OUUW01000001 SPP73853.1 GDIP01025563 JAM78152.1 UFQS01000280 UFQT01000280 SSX02346.1 SSX22721.1 GBBM01004215 JAC31203.1 JO841353 AEO32970.1 AAMC01089184 AAMC01089185 BC075280 AAH75280.1 GDIQ01119055 JAL32671.1 GDIQ01227017 JAK24708.1 GDIQ01206372 JAK45353.1 GAMC01008220 GAMC01008219 JAB98336.1 GFDF01000372 JAV13712.1 GEDC01014505 JAS22793.1 GBXI01015116 GBXI01003618 JAC99175.1 JAD10674.1 CH902619 KPU75734.1 GDIQ01003590 JAN91147.1 GL437132 EFN70946.1 QOIP01000013 RLU15351.1 CH933808 KRG05209.1 KRG05210.1 KRG05211.1 GAKP01007275 GAKP01007274 GAKP01007273 JAC51677.1 CR942545 CAJ81741.1 AK417914 BAN21129.1 JRES01000160 KNC33732.1 CH940648 KRF79928.1 KRF79929.1 KRF79930.1 KRF79931.1 KQ434786 KZC05125.1 GFAC01003696 JAT95492.1 GL732574 EFX75643.1 GEDC01019086 GEDC01014910 JAS18212.1 JAS22388.1 BC041511 CM004478 AAH41511.1 OCT73259.1 QCYY01001749 ROT75645.1 SPP73854.1 EDY69293.2 KRT02805.1 KZ288392 PBC26340.1 CM000158 KRJ99521.1 KQ971338 EFA02967.1 AE013599 BT003193 BT012447 BT033012 AAF58646.3 AAO24948.1 AAS93718.1 ACD99576.1 IACF01001360 LAB67067.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000283053
UP000069940
UP000249989
+ More
UP000008820 UP000076407 UP000075880 UP000075920 UP000075840 UP000007062 UP000075903 UP000075886 UP000000673 UP000075900 UP000075883 UP000075882 UP000069272 UP000030765 UP000075885 UP000002358 UP000075884 UP000027135 UP000009046 UP000076408 UP000078200 UP000075881 UP000183832 UP000095300 UP000002320 UP000095301 UP000008237 UP000005205 UP000235965 UP000091820 UP000001819 UP000268350 UP000008143 UP000007801 UP000075902 UP000000311 UP000279307 UP000009192 UP000037069 UP000008792 UP000076502 UP000000305 UP000186698 UP000283509 UP000242457 UP000005203 UP000002282 UP000007266 UP000092443 UP000000803
UP000008820 UP000076407 UP000075880 UP000075920 UP000075840 UP000007062 UP000075903 UP000075886 UP000000673 UP000075900 UP000075883 UP000075882 UP000069272 UP000030765 UP000075885 UP000002358 UP000075884 UP000027135 UP000009046 UP000076408 UP000078200 UP000075881 UP000183832 UP000095300 UP000002320 UP000095301 UP000008237 UP000005205 UP000235965 UP000091820 UP000001819 UP000268350 UP000008143 UP000007801 UP000075902 UP000000311 UP000279307 UP000009192 UP000037069 UP000008792 UP000076502 UP000000305 UP000186698 UP000283509 UP000242457 UP000005203 UP000002282 UP000007266 UP000092443 UP000000803
Pfam
PF00753 Lactamase_B
Interpro
Gene 3D
ProteinModelPortal
H9JFL9
A0A194PGE0
A0A2A4JYA9
A0A3S2LPT8
A0A2H1WFB4
A0A023EMQ1
+ More
Q16V29 Q1HR84 A0A182XKE3 A0A182JJL5 A0A182VXS1 A0A182HJK1 Q7PPW0 A0A1Q3FYK0 A0A182VMD3 A0A2M3ZKV2 A0A1S4FN35 A0A182QI82 W5JLL9 A0A182RQQ5 A0A182MN27 A0A2M3ZBV3 A0A2M4BUI4 A0A182KWQ8 A0A2M4BUN5 A0A182FP83 A0A084VC25 A0A2M4ALW4 A0A182P8P6 A0A2M4ALV5 A0A2M4ALB8 K7J5S8 A0A336MVK9 A0A182NFU4 A0A067QTI4 A0A336MQI3 E0W3Y8 D3TQQ7 A0A182XXA9 A0A1A9UIZ4 A0A182K9S8 A0A1J1HFT1 A0A1I8PJE4 B0XDM9 A0A1B6FJV1 T1PER7 A0A1B6HTX8 A0A1I8NHM9 E2C1H5 A0A1Y1N6W1 A0A2M4AN96 A0A0N7ZC78 A0A158NIL8 A0A2J7PGF2 A0A1A9WP43 A0A1L8E4Q3 A0A0P5WQ95 R4UMF5 A0A0P5HIF7 N6WCA6 A0A3B0J3J3 A0A0P6BS76 A0A336M0A0 A0A023GE00 G3MHK2 Q6DJA2 A0A0P5QCN0 A0A0P5HGY7 A0A0P5J5A2 W8BYG1 A0A1L8E5A6 A0A1B6DAS7 A0A0A1WKG7 A0A0P8XM15 A0A0P6IJ81 A0A182U7B4 E2A6I9 A0A3L8D5C5 A0A0Q9XC68 A0A034WBL6 Q28BZ5 R4WDW4 A0A0L0CND8 A0A0Q9W4E1 A0A154NZN0 A0A1E1X8A1 E9GY10 A0A1B6D9M4 Q8AVQ9 A0A3R7QRL1 A0A3B0IZL4 B5E059 A0A2A3E3R6 A0A088AAM8 A0A0R1DWE9 D2A1Y2 A0A1A9YPH6 Q86PD3 A0A2P2HZ67
Q16V29 Q1HR84 A0A182XKE3 A0A182JJL5 A0A182VXS1 A0A182HJK1 Q7PPW0 A0A1Q3FYK0 A0A182VMD3 A0A2M3ZKV2 A0A1S4FN35 A0A182QI82 W5JLL9 A0A182RQQ5 A0A182MN27 A0A2M3ZBV3 A0A2M4BUI4 A0A182KWQ8 A0A2M4BUN5 A0A182FP83 A0A084VC25 A0A2M4ALW4 A0A182P8P6 A0A2M4ALV5 A0A2M4ALB8 K7J5S8 A0A336MVK9 A0A182NFU4 A0A067QTI4 A0A336MQI3 E0W3Y8 D3TQQ7 A0A182XXA9 A0A1A9UIZ4 A0A182K9S8 A0A1J1HFT1 A0A1I8PJE4 B0XDM9 A0A1B6FJV1 T1PER7 A0A1B6HTX8 A0A1I8NHM9 E2C1H5 A0A1Y1N6W1 A0A2M4AN96 A0A0N7ZC78 A0A158NIL8 A0A2J7PGF2 A0A1A9WP43 A0A1L8E4Q3 A0A0P5WQ95 R4UMF5 A0A0P5HIF7 N6WCA6 A0A3B0J3J3 A0A0P6BS76 A0A336M0A0 A0A023GE00 G3MHK2 Q6DJA2 A0A0P5QCN0 A0A0P5HGY7 A0A0P5J5A2 W8BYG1 A0A1L8E5A6 A0A1B6DAS7 A0A0A1WKG7 A0A0P8XM15 A0A0P6IJ81 A0A182U7B4 E2A6I9 A0A3L8D5C5 A0A0Q9XC68 A0A034WBL6 Q28BZ5 R4WDW4 A0A0L0CND8 A0A0Q9W4E1 A0A154NZN0 A0A1E1X8A1 E9GY10 A0A1B6D9M4 Q8AVQ9 A0A3R7QRL1 A0A3B0IZL4 B5E059 A0A2A3E3R6 A0A088AAM8 A0A0R1DWE9 D2A1Y2 A0A1A9YPH6 Q86PD3 A0A2P2HZ67
PDB
4CHL
E-value=5.39385e-83,
Score=782
Ontologies
PATHWAY
GO
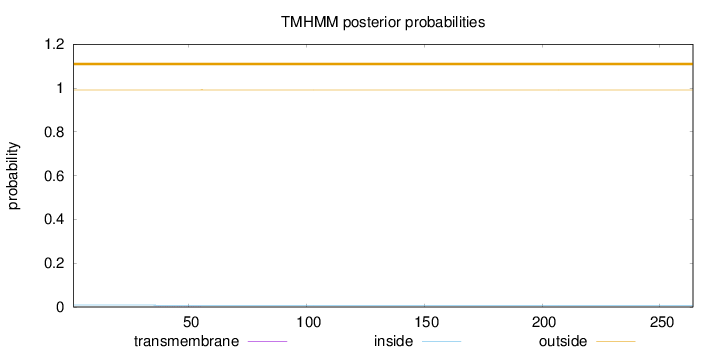
Topology
Length:
264
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00708999999999999
Exp number, first 60 AAs:
0.00691
Total prob of N-in:
0.00870
outside
1 - 264
Population Genetic Test Statistics
Pi
178.632995
Theta
155.405283
Tajima's D
0.626148
CLR
0.012628
CSRT
0.551072446377681
Interpretation
Uncertain
