Gene
KWMTBOMO11051 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008542
Annotation
vacuolar_ATP_synthase_subunit_D_[Bombyx_mori]
Full name
V-type proton ATPase subunit D
+ More
V-type proton ATPase subunit D 1
V-type proton ATPase subunit D 1
Alternative Name
Vacuolar proton pump subunit D
Vacuolar H+ ATPase subunit 36-1
Vacuolar proton pump subunit D 1
dV-ATPase D
Vacuolar H+ ATPase subunit 36-1
Vacuolar proton pump subunit D 1
dV-ATPase D
Location in the cell
Cytoplasmic Reliability : 1.169 Mitochondrial Reliability : 1.625
Sequence
CDS
ATGTCTGGGAAAGATAGATTAGCGATTTTCCCTTCTCGGGGTGCCCAGATGTTAATTAAGGGTCGCCTGGCTGGTGCAGTGAAAGGCCATGGTCTCCTCAAGAAGAAGGCTGATGCCCTTCAAGTGAGGTTCCGTATGATCTTGAGCAAAATCATTGAGACTAAAACCCTTATGGGTGAAGTGATGAAAGAAGCTGCTTTCTCTTTGGCTGAAGCTAAGTTCACAACTGGAGACTTCAACCAAGTTGTCCTACAAAATGTTACCAAGGCTCAAATCAAGATTAGGTCCAAGAAGGACAATGTTGCTGGTGTCACCCTCCCAATCTTTGAGTCATACCAGGATGGTTCTGATACCTACGAGTTGGCTGGTTTGGCCCGTGGTGGGCAGCAGCTTGCAAAGCTGAAGAAGAACTTCCAGAGCGCTGTAAAGCTTTTGGTCGAGTTAGCTTCACTGCAGACTTCATTTGTCACTCTCGACGAGGTTATTAAGATCACGAATAGACGTGTCAACGCCATTGAGCACGTAATCATTCCCCGGCTGGAACGTACACTCGCGTACATCATCTCCGAGCTGGACGAGCTTGAGCGTGAGGAGTTCTACCGTCTCAAGAAGATCCAGGACAAGAAGAAGATTATCAAGGACAAGGCAGAAGCGAAAAAAGCCGCTCTCCTGGCTGCCGGCAACGACTTGCGCGGTGGGGTCACCAACTTGCTGGACGAGGGCGATGAGGACTTGCTGTTCTAA
Protein
MSGKDRLAIFPSRGAQMLIKGRLAGAVKGHGLLKKKADALQVRFRMILSKIIETKTLMGEVMKEAAFSLAEAKFTTGDFNQVVLQNVTKAQIKIRSKKDNVAGVTLPIFESYQDGSDTYELAGLARGGQQLAKLKKNFQSAVKLLVELASLQTSFVTLDEVIKITNRRVNAIEHVIIPRLERTLAYIISELDELEREEFYRLKKIQDKKKIIKDKAEAKKAALLAAGNDLRGGVTNLLDEGDEDLLF
Summary
Description
Subunit of the peripheral V1 complex of vacuolar ATPase. V-ATPase is responsible for acidifying a variety of intracellular compartments in eukaryotic cells, thus providing most of the energy required for transport processes in the vacuolar system (By similarity).
Subunit
V-ATPase is a heteromultimeric enzyme composed of a peripheral catalytic V1 complex (components A to H) attached to an integral membrane V0 proton pore complex (components: a, c, c', c'' and d).
Similarity
Belongs to the V-ATPase D subunit family.
Keywords
Hydrogen ion transport
Ion transport
Transport
Complete proteome
Reference proteome
Feature
chain V-type proton ATPase subunit D
Uniprot
Q2F5J5
A0A0B5L9U3
A0A0N1PHG0
A0A212EXI0
S4PT98
Q9U0S4
+ More
A0A2A4JDX5 A0A194PGN0 A0A2H1W3N8 A0A1J1IJ98 A0A0R6XLQ4 A0A0L0C3F9 A0A1E1WSZ4 H9JG95 A0A1L8EEW8 A0A1I8Q819 K7J5T1 T1PHY7 A0A1A9XA11 A0A1A9VWQ3 A0A1B0AXZ3 A0A1B0A184 A0A1A9WWX7 D3TLR6 Q6PPH4 A0A1B6I5Z8 A0A0A1XE03 A0A0K8WCV4 A0A034WM46 W8B2U5 A0A0K8TRG0 A0A1I9WLG3 D2A2Z0 B4KNU4 E2BRT1 A0A1B0GMK9 B4NMT5 A0A1B6FD11 E0W3E9 M9TIB0 A0A0M4EJR8 A0A1W4WCQ3 A0A0J9RDK9 B4P7C0 Q9V7D2 B3NQB7 A0A1Y1MDE4 B4J8E1 A0A154PQ92 A0A182PW81 A0A158NZX1 B3MHH8 B4LJW3 A0A1L8DFE3 A0A3B0JSS3 Q28YM9 B4H8F2 A0A026WZR7 A0A182KGI3 A0A084W5Q0 A0A1S4H3M6 A0A182VPT2 A0A182KYE4 A0A182RPK1 A0A0J7L6D8 A0A2M3Z3G3 A0A2M4A4L8 A0A2M4BY29 A0A182QDZ7 W5JTQ6 A0A182YN37 A0A0M8ZT39 A0A182IL65 J3JU11 A0A1B6E9N4 A0A1Q3FYT3 A0A1B0CX38 A0A1W4WCL2 A0A023EL05 A0A182F8V0 Q1HQU5 V5GKJ3 V9IJ71 A0A088AP17 A0A0P4VLF5 R4FQ03 A0A182MBX6 A0A0C9RVM8 A0A336MIU6 A0A0V0G4Y6 A0A2S2QL20 A0A023FBA3 U5EZF8 A6YPI5 A0A2H8TGT0 A0A2J7PW84 B4QGW0 A0A067QTK5 Q201Y0 A0A0C9RZD5
A0A2A4JDX5 A0A194PGN0 A0A2H1W3N8 A0A1J1IJ98 A0A0R6XLQ4 A0A0L0C3F9 A0A1E1WSZ4 H9JG95 A0A1L8EEW8 A0A1I8Q819 K7J5T1 T1PHY7 A0A1A9XA11 A0A1A9VWQ3 A0A1B0AXZ3 A0A1B0A184 A0A1A9WWX7 D3TLR6 Q6PPH4 A0A1B6I5Z8 A0A0A1XE03 A0A0K8WCV4 A0A034WM46 W8B2U5 A0A0K8TRG0 A0A1I9WLG3 D2A2Z0 B4KNU4 E2BRT1 A0A1B0GMK9 B4NMT5 A0A1B6FD11 E0W3E9 M9TIB0 A0A0M4EJR8 A0A1W4WCQ3 A0A0J9RDK9 B4P7C0 Q9V7D2 B3NQB7 A0A1Y1MDE4 B4J8E1 A0A154PQ92 A0A182PW81 A0A158NZX1 B3MHH8 B4LJW3 A0A1L8DFE3 A0A3B0JSS3 Q28YM9 B4H8F2 A0A026WZR7 A0A182KGI3 A0A084W5Q0 A0A1S4H3M6 A0A182VPT2 A0A182KYE4 A0A182RPK1 A0A0J7L6D8 A0A2M3Z3G3 A0A2M4A4L8 A0A2M4BY29 A0A182QDZ7 W5JTQ6 A0A182YN37 A0A0M8ZT39 A0A182IL65 J3JU11 A0A1B6E9N4 A0A1Q3FYT3 A0A1B0CX38 A0A1W4WCL2 A0A023EL05 A0A182F8V0 Q1HQU5 V5GKJ3 V9IJ71 A0A088AP17 A0A0P4VLF5 R4FQ03 A0A182MBX6 A0A0C9RVM8 A0A336MIU6 A0A0V0G4Y6 A0A2S2QL20 A0A023FBA3 U5EZF8 A6YPI5 A0A2H8TGT0 A0A2J7PW84 B4QGW0 A0A067QTK5 Q201Y0 A0A0C9RZD5
Pubmed
26354079
22118469
23622113
11030595
26108605
19121390
+ More
20075255 25315136 20353571 25830018 25348373 24495485 26369729 27538518 18362917 19820115 17994087 20798317 20566863 23950915 22936249 17550304 10731132 12537572 12537569 28004739 21347285 15632085 24508170 30249741 24438588 12364791 20966253 20920257 23761445 25244985 22516182 24945155 26483478 17204158 17510324 27129103 25474469 18207082 24845553
20075255 25315136 20353571 25830018 25348373 24495485 26369729 27538518 18362917 19820115 17994087 20798317 20566863 23950915 22936249 17550304 10731132 12537572 12537569 28004739 21347285 15632085 24508170 30249741 24438588 12364791 20966253 20920257 23761445 25244985 22516182 24945155 26483478 17204158 17510324 27129103 25474469 18207082 24845553
EMBL
DQ311428
ABD36372.1
KM923795
AJG43854.1
KQ460813
KPJ12145.1
+ More
AGBW02011741 OWR46202.1 GAIX01012413 JAA80147.1 AJ251992 NWSH01001778 PCG70171.1 KQ459604 KPI92462.1 ODYU01006106 SOQ47658.1 CVRI01000048 CRK99134.1 KP341656 GDQN01009026 AKK21115.1 JAT82028.1 JRES01000955 KNC26777.1 GDQN01001123 JAT89931.1 BABH01002283 GFDG01001561 JAV17238.1 AAZX01020590 KA647488 AFP62117.1 JXJN01005460 CCAG010000155 EZ422368 ADD18644.1 AY588072 AAT01084.1 GECU01025355 JAS82351.1 GBXI01005100 JAD09192.1 GDHF01003397 JAI48917.1 GAKP01003732 JAC55220.1 GAMC01013683 JAB92872.1 GDAI01000877 JAI16726.1 KU932347 APA33983.1 KQ971338 EFA02241.1 CH933808 EDW08989.1 GL450024 EFN81600.1 AJVK01026270 CH964282 EDW85674.1 GECZ01021682 JAS48087.1 DS235882 EEB20155.1 KC473162 AGJ51962.1 CP012524 ALC41542.1 CM002911 KMY94011.1 CM000158 EDW92065.1 AF218238 AE013599 AY069116 CH954179 EDV55893.1 GEZM01036930 JAV82335.1 CH916367 EDW02300.1 KQ435007 KZC13534.1 ADTU01005077 CH902619 EDV37978.2 CH940648 EDW61617.1 GFDF01008979 JAV05105.1 OUUW01000001 SPP75781.1 CM000071 EAL25936.1 CH479223 EDW34987.1 KK107063 QOIP01000005 EZA61236.1 RLU22589.1 ATLV01020682 KE525304 KFB45544.1 AAAB01008971 LBMM01000532 KMQ98118.1 GGFM01002282 MBW23033.1 GGFK01002435 MBW35756.1 GGFJ01008773 MBW57914.1 AXCN02001878 ADMH02000408 ETN66668.1 KQ435859 KOX70575.1 BT126722 AEE61684.1 GEDC01010438 GEDC01002646 JAS26860.1 JAS34652.1 GFDL01002338 JAV32707.1 AJWK01033182 JXUM01064039 JXUM01064040 GAPW01003726 KQ562279 JAC09872.1 KXJ76246.1 DQ440349 CH477615 ABF18382.1 EAT38272.1 GALX01003877 JAB64589.1 JR048522 AEY60692.1 GDKW01000648 JAI55947.1 GAHY01000649 JAA76861.1 AXCM01002501 GBYB01013015 JAG82782.1 UFQS01001095 UFQT01000769 UFQT01001095 SSX08978.1 SSX27087.1 SSX28889.1 GECL01003002 JAP03122.1 GGMS01009160 MBY78363.1 GBBI01000072 JAC18640.1 GANO01000874 JAB58997.1 EF638996 ABR27881.1 GFXV01001436 MBW13241.1 NEVH01020936 PNF20598.1 CM000362 EDX07223.1 KK852969 KDR13105.1 ABLF02041083 DQ413206 ABD72674.1 GBYB01013491 JAG83258.1
AGBW02011741 OWR46202.1 GAIX01012413 JAA80147.1 AJ251992 NWSH01001778 PCG70171.1 KQ459604 KPI92462.1 ODYU01006106 SOQ47658.1 CVRI01000048 CRK99134.1 KP341656 GDQN01009026 AKK21115.1 JAT82028.1 JRES01000955 KNC26777.1 GDQN01001123 JAT89931.1 BABH01002283 GFDG01001561 JAV17238.1 AAZX01020590 KA647488 AFP62117.1 JXJN01005460 CCAG010000155 EZ422368 ADD18644.1 AY588072 AAT01084.1 GECU01025355 JAS82351.1 GBXI01005100 JAD09192.1 GDHF01003397 JAI48917.1 GAKP01003732 JAC55220.1 GAMC01013683 JAB92872.1 GDAI01000877 JAI16726.1 KU932347 APA33983.1 KQ971338 EFA02241.1 CH933808 EDW08989.1 GL450024 EFN81600.1 AJVK01026270 CH964282 EDW85674.1 GECZ01021682 JAS48087.1 DS235882 EEB20155.1 KC473162 AGJ51962.1 CP012524 ALC41542.1 CM002911 KMY94011.1 CM000158 EDW92065.1 AF218238 AE013599 AY069116 CH954179 EDV55893.1 GEZM01036930 JAV82335.1 CH916367 EDW02300.1 KQ435007 KZC13534.1 ADTU01005077 CH902619 EDV37978.2 CH940648 EDW61617.1 GFDF01008979 JAV05105.1 OUUW01000001 SPP75781.1 CM000071 EAL25936.1 CH479223 EDW34987.1 KK107063 QOIP01000005 EZA61236.1 RLU22589.1 ATLV01020682 KE525304 KFB45544.1 AAAB01008971 LBMM01000532 KMQ98118.1 GGFM01002282 MBW23033.1 GGFK01002435 MBW35756.1 GGFJ01008773 MBW57914.1 AXCN02001878 ADMH02000408 ETN66668.1 KQ435859 KOX70575.1 BT126722 AEE61684.1 GEDC01010438 GEDC01002646 JAS26860.1 JAS34652.1 GFDL01002338 JAV32707.1 AJWK01033182 JXUM01064039 JXUM01064040 GAPW01003726 KQ562279 JAC09872.1 KXJ76246.1 DQ440349 CH477615 ABF18382.1 EAT38272.1 GALX01003877 JAB64589.1 JR048522 AEY60692.1 GDKW01000648 JAI55947.1 GAHY01000649 JAA76861.1 AXCM01002501 GBYB01013015 JAG82782.1 UFQS01001095 UFQT01000769 UFQT01001095 SSX08978.1 SSX27087.1 SSX28889.1 GECL01003002 JAP03122.1 GGMS01009160 MBY78363.1 GBBI01000072 JAC18640.1 GANO01000874 JAB58997.1 EF638996 ABR27881.1 GFXV01001436 MBW13241.1 NEVH01020936 PNF20598.1 CM000362 EDX07223.1 KK852969 KDR13105.1 ABLF02041083 DQ413206 ABD72674.1 GBYB01013491 JAG83258.1
Proteomes
UP000053240
UP000007151
UP000218220
UP000053268
UP000183832
UP000037069
+ More
UP000005204 UP000095300 UP000002358 UP000095301 UP000092443 UP000078200 UP000092460 UP000092445 UP000091820 UP000092444 UP000007266 UP000009192 UP000008237 UP000092462 UP000007798 UP000009046 UP000092553 UP000192221 UP000002282 UP000000803 UP000008711 UP000001070 UP000076502 UP000075885 UP000005205 UP000007801 UP000008792 UP000268350 UP000001819 UP000008744 UP000053097 UP000279307 UP000075881 UP000030765 UP000075920 UP000075882 UP000075900 UP000036403 UP000075886 UP000000673 UP000076408 UP000053105 UP000075880 UP000092461 UP000192223 UP000069940 UP000249989 UP000069272 UP000008820 UP000005203 UP000075883 UP000235965 UP000000304 UP000027135 UP000007819
UP000005204 UP000095300 UP000002358 UP000095301 UP000092443 UP000078200 UP000092460 UP000092445 UP000091820 UP000092444 UP000007266 UP000009192 UP000008237 UP000092462 UP000007798 UP000009046 UP000092553 UP000192221 UP000002282 UP000000803 UP000008711 UP000001070 UP000076502 UP000075885 UP000005205 UP000007801 UP000008792 UP000268350 UP000001819 UP000008744 UP000053097 UP000279307 UP000075881 UP000030765 UP000075920 UP000075882 UP000075900 UP000036403 UP000075886 UP000000673 UP000076408 UP000053105 UP000075880 UP000092461 UP000192223 UP000069940 UP000249989 UP000069272 UP000008820 UP000005203 UP000075883 UP000235965 UP000000304 UP000027135 UP000007819
PRIDE
Pfam
PF01813 ATP-synt_D
Interpro
IPR002699
V_ATPase_D
ProteinModelPortal
Q2F5J5
A0A0B5L9U3
A0A0N1PHG0
A0A212EXI0
S4PT98
Q9U0S4
+ More
A0A2A4JDX5 A0A194PGN0 A0A2H1W3N8 A0A1J1IJ98 A0A0R6XLQ4 A0A0L0C3F9 A0A1E1WSZ4 H9JG95 A0A1L8EEW8 A0A1I8Q819 K7J5T1 T1PHY7 A0A1A9XA11 A0A1A9VWQ3 A0A1B0AXZ3 A0A1B0A184 A0A1A9WWX7 D3TLR6 Q6PPH4 A0A1B6I5Z8 A0A0A1XE03 A0A0K8WCV4 A0A034WM46 W8B2U5 A0A0K8TRG0 A0A1I9WLG3 D2A2Z0 B4KNU4 E2BRT1 A0A1B0GMK9 B4NMT5 A0A1B6FD11 E0W3E9 M9TIB0 A0A0M4EJR8 A0A1W4WCQ3 A0A0J9RDK9 B4P7C0 Q9V7D2 B3NQB7 A0A1Y1MDE4 B4J8E1 A0A154PQ92 A0A182PW81 A0A158NZX1 B3MHH8 B4LJW3 A0A1L8DFE3 A0A3B0JSS3 Q28YM9 B4H8F2 A0A026WZR7 A0A182KGI3 A0A084W5Q0 A0A1S4H3M6 A0A182VPT2 A0A182KYE4 A0A182RPK1 A0A0J7L6D8 A0A2M3Z3G3 A0A2M4A4L8 A0A2M4BY29 A0A182QDZ7 W5JTQ6 A0A182YN37 A0A0M8ZT39 A0A182IL65 J3JU11 A0A1B6E9N4 A0A1Q3FYT3 A0A1B0CX38 A0A1W4WCL2 A0A023EL05 A0A182F8V0 Q1HQU5 V5GKJ3 V9IJ71 A0A088AP17 A0A0P4VLF5 R4FQ03 A0A182MBX6 A0A0C9RVM8 A0A336MIU6 A0A0V0G4Y6 A0A2S2QL20 A0A023FBA3 U5EZF8 A6YPI5 A0A2H8TGT0 A0A2J7PW84 B4QGW0 A0A067QTK5 Q201Y0 A0A0C9RZD5
A0A2A4JDX5 A0A194PGN0 A0A2H1W3N8 A0A1J1IJ98 A0A0R6XLQ4 A0A0L0C3F9 A0A1E1WSZ4 H9JG95 A0A1L8EEW8 A0A1I8Q819 K7J5T1 T1PHY7 A0A1A9XA11 A0A1A9VWQ3 A0A1B0AXZ3 A0A1B0A184 A0A1A9WWX7 D3TLR6 Q6PPH4 A0A1B6I5Z8 A0A0A1XE03 A0A0K8WCV4 A0A034WM46 W8B2U5 A0A0K8TRG0 A0A1I9WLG3 D2A2Z0 B4KNU4 E2BRT1 A0A1B0GMK9 B4NMT5 A0A1B6FD11 E0W3E9 M9TIB0 A0A0M4EJR8 A0A1W4WCQ3 A0A0J9RDK9 B4P7C0 Q9V7D2 B3NQB7 A0A1Y1MDE4 B4J8E1 A0A154PQ92 A0A182PW81 A0A158NZX1 B3MHH8 B4LJW3 A0A1L8DFE3 A0A3B0JSS3 Q28YM9 B4H8F2 A0A026WZR7 A0A182KGI3 A0A084W5Q0 A0A1S4H3M6 A0A182VPT2 A0A182KYE4 A0A182RPK1 A0A0J7L6D8 A0A2M3Z3G3 A0A2M4A4L8 A0A2M4BY29 A0A182QDZ7 W5JTQ6 A0A182YN37 A0A0M8ZT39 A0A182IL65 J3JU11 A0A1B6E9N4 A0A1Q3FYT3 A0A1B0CX38 A0A1W4WCL2 A0A023EL05 A0A182F8V0 Q1HQU5 V5GKJ3 V9IJ71 A0A088AP17 A0A0P4VLF5 R4FQ03 A0A182MBX6 A0A0C9RVM8 A0A336MIU6 A0A0V0G4Y6 A0A2S2QL20 A0A023FBA3 U5EZF8 A6YPI5 A0A2H8TGT0 A0A2J7PW84 B4QGW0 A0A067QTK5 Q201Y0 A0A0C9RZD5
PDB
6O7X
E-value=3.48274e-48,
Score=481
Ontologies
KEGG
PATHWAY
GO
PANTHER
Topology
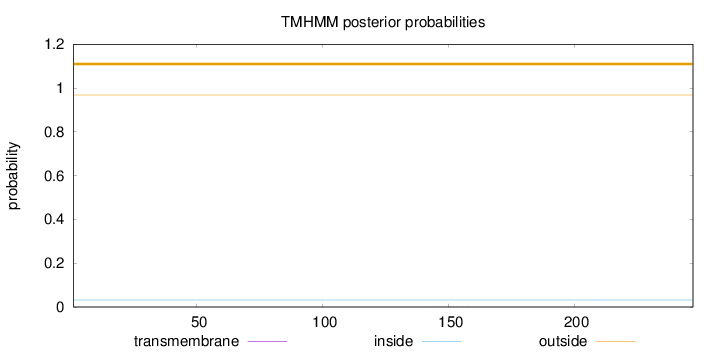
Length:
247
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00462999999999999
Exp number, first 60 AAs:
0.00159
Total prob of N-in:
0.03152
outside
1 - 247
Population Genetic Test Statistics
Pi
18.148157
Theta
164.790074
Tajima's D
0.174877
CLR
1.786311
CSRT
0.418029098545073
Interpretation
Uncertain
