Gene
KWMTBOMO11046
Pre Gene Modal
BGIBMGA008320
Annotation
PREDICTED:_uncharacterized_protein_LOC106136342_isoform_X1_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 3.9
Sequence
CDS
ATGTTATTCTTTTTTGCAGGATGCGCCAACATGTGGCGTTTAGTGCTACTGCTGTGTTTGGCGACTCCAAGTCTCGGGCAGCAGAGTAAGCCCAACAGGGTGGTGAGCGTGGCTACCAAGTGCGACAAGGGTTCCATCATAGTGGACCTGGAGATGGATCGCCCGTTCAAAGGATTAGTATTCAGCAAAGATTTCTCTACTGAATGTCAATCACAAGGCAGTTTAAACACGAAGATATCACTACATCTCCCATCGAACGCGTGCGGTGTGCGAACATCCACATTGAACACCACCGAATTTGAACCAGACGACGAAGAACAATTGTATTATAACGTGGAACTGGTCGTGCAAATGGACAAACACTTGCAACAGTCTTCCGACCAGGAGATCTTGGTCAAATGCAAACTGCAAAGACGAGCAGTCCAAATCGACAGTCCGGCGTTGGAAAGTGTAATCAACAATAAAATAAGGGAAATAGCCGGGCAAGAAATAAAAAAATCCAGAATGGGTCGAAACCGTCAAGGTTTCGCGAGTTCGGCCGAAATGGAGCAGCGCGAAATGCTGATGGAAGCGGCCCGGGCCTGGATGGAATTGTCTCCGGCCGCTGCTGCCACTGAACCGGCCACAGTCGAAGTGGGACAGCCCATGAGATTGCTTATTCAATGTACGCTACCTGTCGGAGTCGGGCTTCGCGTAACAAACTGCCTCGCTCACGATGGTCAAGGCGAAGCCTCACAGAAACTGCTCGACGAAGCCGGCTGTCCGATTGACGAATCAATCTTCAACAAGCCCACCATCCACAGCCACAAGCAGAGGAGCAGCGAAATCGATTTCTCCGAACTCGAACCGGCCGGTACGCAGAGAAATATAGACGAGAGCTTAAGCGTTAAGCAATCGAAGACCGATCCAGATTTTATGTTAAAGAACATAATGACGTACCAACATGCGGTTACGACGTTCGCTGCTTTCAAGTTTCCGGACAGAGCGAAATTACATCTCACCTGTGGCATCGAGTTATGTAAAGGGCAATGCCCTCCTGTGGACTGCAAGGCGTTGCAGAAGCCTCAGCAGACTAGAGATGGATTATTGAGGAAGGCGCGACTCGATAAGGACGCGAGAGGAATCGTCATAGACAGACTGGAAGTGTACAATAGTATCGAGGTTCACGCACCGAATATTGAATTAGAAGATGAAGCTTCTATACGGGGATCGAGAAGGATAGAAGATGAAGAATTTATTCGCGGGTTTTCGCCGGGCGACAAAACGATTTGTCTGTCTCCAGGGAAAATGGCTTTAGCGTTTTGCGTGTTAGGAGTTATTTTCCTTTGTGCCATCGCGGTGGCCTTTGTATCATTGGTGCGAGCGAGACGGCGATCAGGCCGAGAGCCTCTACACACGTCACTATCATTTTATACGGGCAGTAAGAGTATGTTCTCGTCAAGCGGGAGCTCTAGTTCTGGTCTCAGCGGTAGTAAATTGCTGTTAACTGACAGCCCTTACATGGATCACCATTCGTCGTCCAGTAAAAACTGGCCGTATCCCAGGCCATATTGA
Protein
MLFFFAGCANMWRLVLLLCLATPSLGQQSKPNRVVSVATKCDKGSIIVDLEMDRPFKGLVFSKDFSTECQSQGSLNTKISLHLPSNACGVRTSTLNTTEFEPDDEEQLYYNVELVVQMDKHLQQSSDQEILVKCKLQRRAVQIDSPALESVINNKIREIAGQEIKKSRMGRNRQGFASSAEMEQREMLMEAARAWMELSPAAAATEPATVEVGQPMRLLIQCTLPVGVGLRVTNCLAHDGQGEASQKLLDEAGCPIDESIFNKPTIHSHKQRSSEIDFSELEPAGTQRNIDESLSVKQSKTDPDFMLKNIMTYQHAVTTFAAFKFPDRAKLHLTCGIELCKGQCPPVDCKALQKPQQTRDGLLRKARLDKDARGIVIDRLEVYNSIEVHAPNIELEDEASIRGSRRIEDEEFIRGFSPGDKTICLSPGKMALAFCVLGVIFLCAIAVAFVSLVRARRRSGREPLHTSLSFYTGSKSMFSSSGSSSSGLSGSKLLLTDSPYMDHHSSSSKNWPYPRPY
Summary
Uniprot
H9JFM3
A0A2A4JEG7
A0A2H1WSF8
A0A0N0PBS9
A0A194PMW5
A0A212FBD9
+ More
A0A0L7LT82 A0A1Y1N8Q4 D6WTK5 A0A2J7QEK1 A0A1B0EW54 A0A1B0CAP6 A0A0T6BAG5 B4ND11 A0A1I8MML6 B4GXV3 B3MRK5 A0A1I8P6I1 A0A1I8P6J0 B4M1Y9 A0A1B0A182 A0A1W4VI91 B4R6B0 B4Q0M6 A0A336MSG7 Q29HE2 Q86P22 Q9W3K2 B6IDJ0 B4IKW8 B4L8I1 A0A336MM46 A0A0M4EM14 B3NWK1 B4JNR1 A0A2P8YT99 A0A3B0K405 A0A1J1IFY0 A0A0P4W2U0 A0A1D2MF36 A0A0P5JBJ4 A0A1D1UNU0 A0A1Y1MK04 A0A1Y1MG57 A0A2H8TJT0 D6WRI4 A0A1J1IVA0 A0A1B0GP52 J9JLK5 A0A232EVI1 A0A0A9X3T3 A0A1L8DA00 A0A182L393 A0A0K8TK78 Q7QIT1 A0A182II10
A0A0L7LT82 A0A1Y1N8Q4 D6WTK5 A0A2J7QEK1 A0A1B0EW54 A0A1B0CAP6 A0A0T6BAG5 B4ND11 A0A1I8MML6 B4GXV3 B3MRK5 A0A1I8P6I1 A0A1I8P6J0 B4M1Y9 A0A1B0A182 A0A1W4VI91 B4R6B0 B4Q0M6 A0A336MSG7 Q29HE2 Q86P22 Q9W3K2 B6IDJ0 B4IKW8 B4L8I1 A0A336MM46 A0A0M4EM14 B3NWK1 B4JNR1 A0A2P8YT99 A0A3B0K405 A0A1J1IFY0 A0A0P4W2U0 A0A1D2MF36 A0A0P5JBJ4 A0A1D1UNU0 A0A1Y1MK04 A0A1Y1MG57 A0A2H8TJT0 D6WRI4 A0A1J1IVA0 A0A1B0GP52 J9JLK5 A0A232EVI1 A0A0A9X3T3 A0A1L8DA00 A0A182L393 A0A0K8TK78 Q7QIT1 A0A182II10
Pubmed
EMBL
BABH01002273
BABH01002274
NWSH01001778
PCG70166.1
ODYU01010700
SOQ55978.1
+ More
KQ460813 KPJ12142.1 KQ459604 KPI92465.1 AGBW02009352 OWR51033.1 JTDY01000147 KOB78599.1 GEZM01009877 JAV94253.1 KQ971354 EFA07190.1 NEVH01015308 PNF27014.1 AJVK01026270 AJWK01004149 LJIG01002753 KRT84187.1 CH964239 EDW82720.2 CH479196 EDW27580.1 CH902622 EDV34410.1 CH940651 EDW65693.1 CM000366 EDX17387.1 CM000162 EDX02297.1 UFQS01001095 UFQT01001095 SSX08977.1 SSX28888.1 CH379064 EAL31816.2 KRT06592.1 KRT06593.1 BT003525 AAO39529.1 AE014298 BT133192 AAF46324.2 AFA28433.1 AHN59475.1 BT050430 ACJ13137.1 CH480859 EDW52739.1 CH933815 EDW07956.1 UFQT01000769 SSX27088.1 CP012528 ALC48658.1 CH954180 EDV46821.2 CH916371 EDV92354.1 PYGN01000374 PSN47471.1 OUUW01000016 SPP88895.1 CVRI01000048 CRK99128.1 GDRN01076966 JAI62810.1 LJIJ01001480 ODM91610.1 GDIQ01208106 JAK43619.1 BDGG01000001 GAU88947.1 GEZM01032293 JAV84685.1 GEZM01032294 JAV84684.1 GFXV01001733 MBW13538.1 KQ971351 EFA06573.1 CVRI01000059 CRL03070.1 AJVK01033134 AJVK01033135 ABLF02041159 NNAY01001991 OXU22366.1 GBHO01028212 JAG15392.1 GFDF01010940 JAV03144.1 GDAI01002926 JAI14677.1 AAAB01008807 EAA03968.4 APCN01001489
KQ460813 KPJ12142.1 KQ459604 KPI92465.1 AGBW02009352 OWR51033.1 JTDY01000147 KOB78599.1 GEZM01009877 JAV94253.1 KQ971354 EFA07190.1 NEVH01015308 PNF27014.1 AJVK01026270 AJWK01004149 LJIG01002753 KRT84187.1 CH964239 EDW82720.2 CH479196 EDW27580.1 CH902622 EDV34410.1 CH940651 EDW65693.1 CM000366 EDX17387.1 CM000162 EDX02297.1 UFQS01001095 UFQT01001095 SSX08977.1 SSX28888.1 CH379064 EAL31816.2 KRT06592.1 KRT06593.1 BT003525 AAO39529.1 AE014298 BT133192 AAF46324.2 AFA28433.1 AHN59475.1 BT050430 ACJ13137.1 CH480859 EDW52739.1 CH933815 EDW07956.1 UFQT01000769 SSX27088.1 CP012528 ALC48658.1 CH954180 EDV46821.2 CH916371 EDV92354.1 PYGN01000374 PSN47471.1 OUUW01000016 SPP88895.1 CVRI01000048 CRK99128.1 GDRN01076966 JAI62810.1 LJIJ01001480 ODM91610.1 GDIQ01208106 JAK43619.1 BDGG01000001 GAU88947.1 GEZM01032293 JAV84685.1 GEZM01032294 JAV84684.1 GFXV01001733 MBW13538.1 KQ971351 EFA06573.1 CVRI01000059 CRL03070.1 AJVK01033134 AJVK01033135 ABLF02041159 NNAY01001991 OXU22366.1 GBHO01028212 JAG15392.1 GFDF01010940 JAV03144.1 GDAI01002926 JAI14677.1 AAAB01008807 EAA03968.4 APCN01001489
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
+ More
UP000007266 UP000235965 UP000092462 UP000092461 UP000007798 UP000095301 UP000008744 UP000007801 UP000095300 UP000008792 UP000092445 UP000192221 UP000000304 UP000002282 UP000001819 UP000000803 UP000001292 UP000009192 UP000092553 UP000008711 UP000001070 UP000245037 UP000268350 UP000183832 UP000094527 UP000186922 UP000007819 UP000215335 UP000075882 UP000007062 UP000075840
UP000007266 UP000235965 UP000092462 UP000092461 UP000007798 UP000095301 UP000008744 UP000007801 UP000095300 UP000008792 UP000092445 UP000192221 UP000000304 UP000002282 UP000001819 UP000000803 UP000001292 UP000009192 UP000092553 UP000008711 UP000001070 UP000245037 UP000268350 UP000183832 UP000094527 UP000186922 UP000007819 UP000215335 UP000075882 UP000007062 UP000075840
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JFM3
A0A2A4JEG7
A0A2H1WSF8
A0A0N0PBS9
A0A194PMW5
A0A212FBD9
+ More
A0A0L7LT82 A0A1Y1N8Q4 D6WTK5 A0A2J7QEK1 A0A1B0EW54 A0A1B0CAP6 A0A0T6BAG5 B4ND11 A0A1I8MML6 B4GXV3 B3MRK5 A0A1I8P6I1 A0A1I8P6J0 B4M1Y9 A0A1B0A182 A0A1W4VI91 B4R6B0 B4Q0M6 A0A336MSG7 Q29HE2 Q86P22 Q9W3K2 B6IDJ0 B4IKW8 B4L8I1 A0A336MM46 A0A0M4EM14 B3NWK1 B4JNR1 A0A2P8YT99 A0A3B0K405 A0A1J1IFY0 A0A0P4W2U0 A0A1D2MF36 A0A0P5JBJ4 A0A1D1UNU0 A0A1Y1MK04 A0A1Y1MG57 A0A2H8TJT0 D6WRI4 A0A1J1IVA0 A0A1B0GP52 J9JLK5 A0A232EVI1 A0A0A9X3T3 A0A1L8DA00 A0A182L393 A0A0K8TK78 Q7QIT1 A0A182II10
A0A0L7LT82 A0A1Y1N8Q4 D6WTK5 A0A2J7QEK1 A0A1B0EW54 A0A1B0CAP6 A0A0T6BAG5 B4ND11 A0A1I8MML6 B4GXV3 B3MRK5 A0A1I8P6I1 A0A1I8P6J0 B4M1Y9 A0A1B0A182 A0A1W4VI91 B4R6B0 B4Q0M6 A0A336MSG7 Q29HE2 Q86P22 Q9W3K2 B6IDJ0 B4IKW8 B4L8I1 A0A336MM46 A0A0M4EM14 B3NWK1 B4JNR1 A0A2P8YT99 A0A3B0K405 A0A1J1IFY0 A0A0P4W2U0 A0A1D2MF36 A0A0P5JBJ4 A0A1D1UNU0 A0A1Y1MK04 A0A1Y1MG57 A0A2H8TJT0 D6WRI4 A0A1J1IVA0 A0A1B0GP52 J9JLK5 A0A232EVI1 A0A0A9X3T3 A0A1L8DA00 A0A182L393 A0A0K8TK78 Q7QIT1 A0A182II10
Ontologies
Topology
SignalP
Position: 1 - 26,
Likelihood: 0.992814
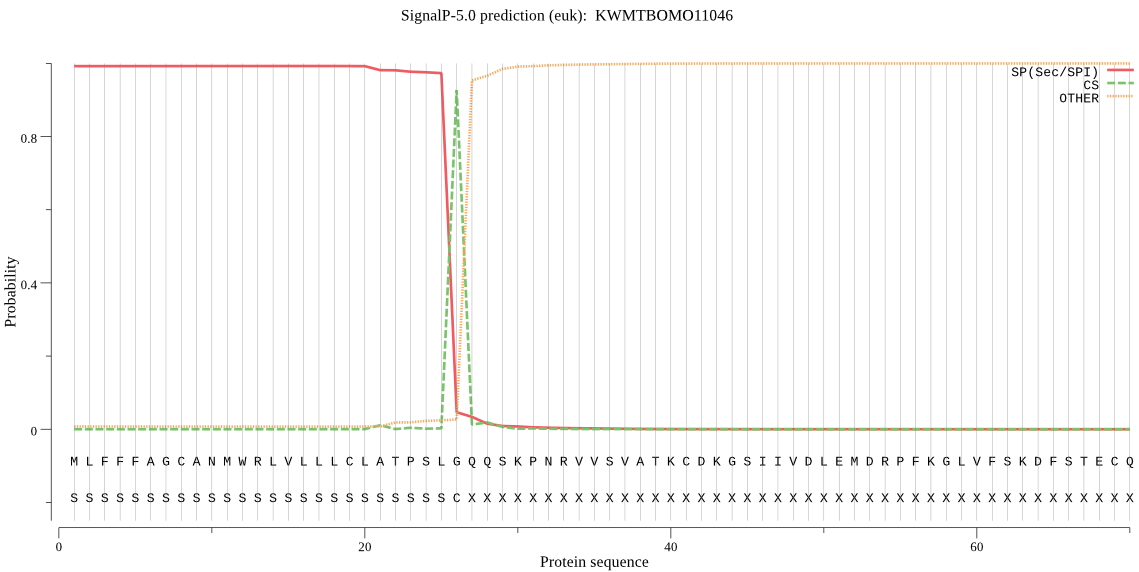
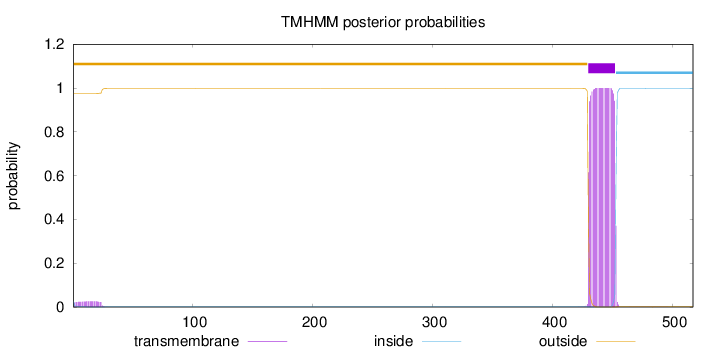
Length:
517
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.4062
Exp number, first 60 AAs:
0.57055
Total prob of N-in:
0.02612
outside
1 - 429
TMhelix
430 - 452
inside
453 - 517
Population Genetic Test Statistics
Pi
278.283642
Theta
179.083532
Tajima's D
0.759789
CLR
1.117133
CSRT
0.598870056497175
Interpretation
Uncertain
