Gene
KWMTBOMO11045 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008321
Annotation
PREDICTED:_spectrin_beta_chain_isoform_X2_[Amyelois_transitella]
Full name
Spectrin beta chain
Location in the cell
Cytoplasmic Reliability : 1.668 Nuclear Reliability : 1.397
Sequence
CDS
ATGACGACCGACATCTCGGTGTCCCGATGGGACCCGTCCGTCGGCCACGGACAGGAGATCATTGATGAAATCGAGTACGACGGCGGAAATTCATCGTCGAGGTTATTCGAACGATCGCGCATTAAAGCGCTTGCAGACGAACGAGAGAGCGTGCAAAAGAAGACATTTCAAAAATGGGTGAACTCACATTTAGTTCGCGTCGGGTTCCGAATTGGGGATCTGTACGTCGATATGAGAGATGGAAAGATGCTTATCAGACTACTCGAGGTGCTGTCTGGAGAGAGACTGCCTCGTCCAACAAAAGGCAAAATGCGTATCCACTGCTTGGAGAACGTGGACAAGGCGCTTCAGTTTCTGCGCGAACAAAGAGTGCACCTGGAGAACATGGGGTCCCATGACATCGTCGACGGCAACCCTAGGCTCAGTCTTGGTCTCATCTGGACTATCATTCTGCGCTTCCAGATCCAAGACATTACAATCGAAGAAACCGACAACAAGGAAACGAAATCGGCGAAGGACGCTCTCCTTCTTTGGTGTCAAATGAAGACGGCGGGTTATAACAACGTGAACGTCCGCAACTTCACGACGTCCTGGCGGGACGGGTTGGCGTTCAACGCGATCATACACAAGCACAGGCCCGACCTTGTTCAGTTCGAGAAACTGCACAGGAGCAACCCTATCCACAACCTTAACAACGCCTTCAACGTTGCCGAGGAAAAGCTTGGACTCACCAAGTTGCTTGACGCGGAAGACATCGCTGTCGACCATCCCGACGAGAAGTCCATCATCACGTACGTGGTGACGTACTATCACTACTTCAGCAAGATGAAGCAGGAGACAGTCCAAGGCAAGAGGATCGGAAAGGTCGTCGGTATCGCGATGGAAAACGACAAGATGATGCAGGAATACGAGTCGCTCACCAGCGATCTCCTGCAGTGGATCGAGCGAACAATCGAGGCGCTCGGCGACAGAACATTCGCGAACTCTCTGGAGGGAGTGCGTCAACAATTGATTCAGTTCGCGAACTATCGCACCGTCGAGAAACCGCCTAAGTTCGTGGAGAAGGGCAACCTGGAGGTGCTGCTGTTCACGCTGCAGTCGCGCATGCGCGCCGCCAACCAGCGCCCGTACACGCCGCGCGAGGGCCGCATGATCCACGACATCAACCGCGCCTGGGAGACGCTGGAGAAGGCCGAGCACGAGCGGGAGCTGGCGCTGCGGGAGGAGCTCATCCGCCAGGAGAAGCTCGAGCAGCTCGCCGCCAGATTCAACCGTAAAGCTCAGATGCGCGAGACCTGGCTGTCGGAGAACCAGCGCCTGGTGAGCCAGGACAACTTCGGGTTCGACCTGGCGGCCGTGGAAGCCGCCGCCAAGAAGCACGAGGCCATCGAGACCGACATCTTCGCGTACGAGGAGCGCGTGCAGGCCGTCGTCGCCGTCTGCTCGGAGCTGCAGGCCGAGAGGTACCACGACATCGAGCGTGTGTGCGCGCGGCGCGACAACGTGCTGCGCCTGTGGGCCTACCTGCTGGAGCTGCTGCGGGCGCGCCGGGCTCGACTCGAACTGTCCCTGCAGCTGCAACAGAACTTCCAGGAAATGCTGTACATCCTCGACTCGATGGAAGAGATCAAGATGCGTCTCTTGACAGACGACTACGGCAAACATTTGATGGGAGTCGAAGATTTATTACAAAAGCATGCGCTGGTTGAAGCCGACATCAACGTACTCGGAGAACGAGTCAAGGTGGTGGTCGGACAGTCGCAGCGCTTCCTGGAGGGCGAGGGCGGCGCGTACCGGCCGTGCGACCCCGCCGTCACCGTGGACCGCATACAGCAGCTCGAGAACGCGTACGCAGAGCTCGTGCACCTCGCCGTGGAGCGCAGAAAGCGTTTGGAGGACAGCCGCAAACTGTGGCAGTTCTACTGGGACATGGCTGATGAAGAGAACTGGATCAAGGAGAAGGAACAGATCGTCTCCGTTGATGACATCGGACACGACCTCACTACCGTGTACTTGCTGATATCGAAGCACAAGGCGCTGGAGGCCGACGTGCTGGCCCACGAGCCGCAGCTAATGAGCGTGGCGGCGGTCGGCGACGAGCTCATCTCCCAGGGACACTTCGGCGCTGACAGGATCCAAGAACGCCTCCGTGACATCCTGTCGCAATGGAACCACCTCCTCGACTTGGTGTCGCTCCGCAAGAGTCGGCTGGACGCCGCCGTGCGCTACCACCAACTGTTTGCTGATGCCGACGACATCGACAACTGGATGCTCGACACCCTCAGATTGGTTTCGTCTGAGGACACCGGCGTTGACGAAGCTCAGGTGCAGAGCCTGCTGAAGAAACATAAGGACGTAACCGACGAACTCAAGAACTACGCCAACGTCATTCAGCAACTGAAGCAACAGGCGCAGGAGCTGAGCCCGGAGGACGCCAACAGCGGCGAGGTGCGCGAGCGGCTGGCGGCCATCGACGCGCGGCACGGCGAGCTGGCCGAGCTGGCGCGCCTCCGCAAGCAGCGCCTGCTGGACGCGCTGTCGCTGTTCAAGCTGCTCGCCGAGGCCGACGCCGCCGACCAGTGGATCGCGGAGAAGCACCGCATGCTCGACACCATGCTGCCGCCCAGCGACATCGACGACGTCGAGATCATGAAGCACAGATACGACGGCTTCGACAAGGAGATGAACGCGAACGCGTCCCGCGTGGCCGTCGTGAACCAGCTGGCGCGGCAGCTCGTGCACGTGGAGCACCCGCAGGCGCCGCGCATCCAGGACCGCCAGCACCAGCTCAACCACGCCTGGAGCGCGCTCCGGGAGAAGGCTGAAGCGAAGAAGGATGAGCTTAAGTCCGCTCAAGGTGTGCAGACATTCCACATCGAATGTCGCGAGACCGTCTCGTGGATCGAGGACAAGAAACGCATACTGCAGCAAACCGACGACCTCGAGATGGATCTTAACGGCGTGATGACTCTCCAACGTCGTTTGTCCGGAATGGAGCGCGACTTGGCCGCCATACAGGCTCGCATCTCGTCCCTGGAGAGCGAGGCCAGCGCCATCGAGAACGAACATCCCGAGGAGGCACAACTCATTCGGGAACGCGTCGAACAGATCACTCAGAACTGGGAGCAACTCACTCACATGCTGAAAGAACGCGACTCGAAATTGGAAGAGGCCGGAGACTTACATCGTTTCCTGCGCTCCGTCGATCATTTCCAGGCGTGGCTCACAAAGACGCAGACCGATGTTGCTTCCGAAGATATCCCATCAAGTCTGCCTGAGGCTGAGAAACTCCTTTCTCAGCACCAAACCATCAAAGAGGAGATTGATAACTATAAGGATGAATATGCTAAGATGATGGAATATGGAGAGAAGATTACTGCCGAACCGTCCACCCAAGATGATCCACAGTATATGTTCCTTCGCGAACGTTTGAAGGCACTGCGCGAGGGATGGGCCGAGCTGCAGCAGATGTGGGAGAACCGACAGCAGCTCCTGGCGCAGAGCCTCGAGTTGCAGCTATTGCAGCGTGATGCCAGACAGGCTGAAGTTTTACTTGCACACCAAGAACACAGGCTCGCCAAAACCGAGCCTCCGACAAACTTGGAACAGGCTGAAAATATTATCAAGGAACACGAGGCCTTCCTCACTACGATGGAGGCTAATGACGATAAGATTAATTCAGTTGTGCAGTTCGCTAATCGCCTCGTCGAGGAGAGGCACTTCGACGCGGACAAGATTCAGCGCAAGGCGGAGAACATCGAGGCACGCCGCAATGCCAACCGCGAAAAGGCACTTAACCTCATGGAGAAGCTCCAAGACCAGCTTCAGTTGCACCAGTTCTTGCAGGACTGCGACGAACTTGGAGAATGGGTTCAGGAGAAGAACGTTACCGCACAAGACGATACCTACCGCTCCGCCAAGACTATCCACTCTAAGTGGACCCGTCATCAGGCTTTCGAAGCTGAAATTGCGGCCAACAAAGAACGACTGTTCGCTGTGCAGAACGCCGCTGAAGAGCTCATGAAGCAGAAACCGGAATTCGTTGAAGTAATTTCGCCCAAGATGCACGAACTCCAAGATCAATTTGAAAACTTGCAGACTACCACCAAGGAGAAGGGCGAGAGGCTCTTCGACGCCAACAGGGAAGTGCTGCTTCACCAAACCTGCGACGACATCGACTCATGGATGAACGAACTTGAGAAGCAAATTCAGAGCACCGATACCGGTAGCGATCTGGCCTCCGTCAATATTCTCATGCAGAAGCAACAGATGATCGAGACCCAGATGGCAGTGAAGGCGAAACAGGTTACGGAACTGGAAACTCAGGCGGAATACCTGCAGAAGACCGTGCCTGACAAGATGGAAGAGATCAAGGAGAAGAAGAAGACTGTTGAGCATAGATTCGAACAACTCAAGGCGCCATTACTCGAACGCCAACGTCACCTTGCCAAGAAGAAGGAAGCCTTCCAGTTCCGTCGTGATGTCGAAGACGAGAAGCTGTGGGTCCAGGAGAAGATGCCCCTGGCCACAAGCACCGACTACGGTAACTCATTGTTCAACGTGCAAATGTTGCAGAAAAAGACTCAGTCGCTGAAGACGGAGACCGACAACCACGAGCCCAGGATTCTGACGGTCATCTCTAACGGACAGAAGCTGATTGACGAGGGACACGAAAGTTCGCCCGAATTCAAACAGCTTATTGACGAGCTCACTGCGAGGTGGCGAGAACTTAAGGATGCCATCGAGGAACGCAAGAACAATCTGTCGCAATGGGAGAAAGCACACCAGTACCTGTTCGATGCGAACGAAGCGGAAGCCTGGATGAGCGAACAGGAACTCTACATGATGGTGGAAGATCGAGGCAAGGATGAGATCTCTGCGCAGAACCTCATGAAGAAACACGAGATTTTGGAGCAAGCTGTCGACGATTATGCACAGACTATCCGTCAATTGGGAGAGACTGTCAGGCAGCTCACCGCCGAAGAACATCCACTCAGTGAACAAATTTCCGTGAAACAATCGCAAGTAGACAAGTTGTATGCAGGACTGAAGGACTTGGCTGGCGAGAGACGAGCCAAACTCGATGAAGCTTTAAGATTATTCCAACTATCCCGCGAGGTCGATGACCTTGAACAATGGATCACAGAGAGGGAACTCGTCGCCAGCAGCCAGGAGCTGGGACAAGACTACGATCATGTCACGCTGCTGTGGGAACGCTTCAAGGAGTTCGCGCGCGAGACCCAGTCGGTGGGCTCGGAGCGAGTCGGCACGGCCGAGCGTATCGCCGACCAGATGATCGCCATGGGACATTCTGACAACGCCACCATCGCGCAGTGGAAGGAGGGCCTCAAGGAAACCTGGCAGGACCTCCTCGAGCTCATCGAGACGAGGACACAGATGTTAGCGGCGTCCCGCGAGCTGCACAAGTACTTCCACGACTGCAAGGACACGCTGCAGCGCGTCAACGAGCGCGCGCGCGGCGTCAGCGACGAGCTCGGCCGCGACGCCATCAGCGTGTCCGCGCTGCAGCGCAAGCACCACAACTTCATGCAGGACCTCACCACGCTGCAGCAGCAGGTGGAGTCCATAAAGAGCGAGTGCTCGCGGCTGGGCGCGTCGTACGCGGGCGAGAAGGCGGCGGAGATCACGCGGCGCGAGCGCGAGGTGGCGGACGCGTGGGCCGCGCTGCAGGCCGCGTGCCGCGCGCGCCGCGCCAAGCTGGAGGACGCCGACCGCCTGTACCGCTTCCTCAGCCAGGTCCGCACGCTCACGCTCTGGATGGACGACGTCGTGCGACAGATGAACACCGGCGAGAAGCCCAGAGACGTGAGCGGCGTCGAGCTTCTGATGAACAACCACCAGTCGCTGAAGGCGGAGATCGACACGCGCGAGGACAACTTCTCGGCCTGCATCTCGCTCGGGAAGGAGCTGCTGGCGCGCCAACACTACGCCTCCGCGGACATCAAGGAGAAGCTACTGCAGCTGACGAATCAAAGGAATGCCCTGCTGCGACGCTGGGAAGAACGCTGGGAGAACTTGCAACTCATACTGGAAGTGTACCAGTTCGCTCGCGACGCGGCCGTGGCGGAGGCGTGGCTCATCGCGCAGGAGCCGTACCTCATGTCGCAGGAGCTGGGGCACACCATCGACGAGGTCGAGAGCCTCATCAAGAAGCACGAGGCCTTCGAGAAATCCGCAGCGGCACAGGAGGACCGGTTCAGCGCTCTGCAGCGACTCACCACCTTCGAAGTGAAAGAGATGAAGCGTCGTCAGGAAGCGGCCGAAGCGGCGGAGCGCGAGAAGCGGGAGCGCGAAGCCGCCGAGGCCGCCGCCGCCGCCGCCGCCGCGCAGGACGCGCTCGACCGCGCCGACACGCCGCACGAGCAGCCCGCCGCCAACGAGGCGAGCTCCGTCCCGAACCCCGCCGCCCTCGCCGCCCCCGCTGACCCCGCCGCCCCCGCACCGCCCTCCCCCCAACAGTCCACGTCGAAACCAACACCGGTTAAACCCGCACGCCTCTCCAAGGGAGCCAGTCCAGCCAGCGAAGATGAAGGGGTCGAGGGAACTCTGGTACGGAAACACGAATGGGAATCCGCCGCCAAACGGGCTTCTAATCGGTCTTGGGATAAGCTTTACGTTGTGGCCAAAGACGGCCGCATGTCGTTCTACAAGGACCAGAAGAGCTACAAGTCGTCGCCGGAACAGTACTGGCGCGGGGAGAGCCCTCTGGACCTCAACGGGGCGGTCGTAGAGGTGGCAGCCAACTACACCAAGAAGAAGCACGTCTTCCGTCTCAGGCTGTCGAGCGGCGCGGAGTTCCTGCTGCAGGCGCACGACGAGGCGGAGATGTCGTGGTGGCTGGAGTCGCTGCGGGCGCGCACGCAGGCGCCGGCCGCGCGCTCGCACACGCTGCCCGCGCCCGCCAACGCCGAGCCCAAGCGACGCTCCTTCTTCACGCTCAAGAAGAACTAA
Protein
MTTDISVSRWDPSVGHGQEIIDEIEYDGGNSSSRLFERSRIKALADERESVQKKTFQKWVNSHLVRVGFRIGDLYVDMRDGKMLIRLLEVLSGERLPRPTKGKMRIHCLENVDKALQFLREQRVHLENMGSHDIVDGNPRLSLGLIWTIILRFQIQDITIEETDNKETKSAKDALLLWCQMKTAGYNNVNVRNFTTSWRDGLAFNAIIHKHRPDLVQFEKLHRSNPIHNLNNAFNVAEEKLGLTKLLDAEDIAVDHPDEKSIITYVVTYYHYFSKMKQETVQGKRIGKVVGIAMENDKMMQEYESLTSDLLQWIERTIEALGDRTFANSLEGVRQQLIQFANYRTVEKPPKFVEKGNLEVLLFTLQSRMRAANQRPYTPREGRMIHDINRAWETLEKAEHERELALREELIRQEKLEQLAARFNRKAQMRETWLSENQRLVSQDNFGFDLAAVEAAAKKHEAIETDIFAYEERVQAVVAVCSELQAERYHDIERVCARRDNVLRLWAYLLELLRARRARLELSLQLQQNFQEMLYILDSMEEIKMRLLTDDYGKHLMGVEDLLQKHALVEADINVLGERVKVVVGQSQRFLEGEGGAYRPCDPAVTVDRIQQLENAYAELVHLAVERRKRLEDSRKLWQFYWDMADEENWIKEKEQIVSVDDIGHDLTTVYLLISKHKALEADVLAHEPQLMSVAAVGDELISQGHFGADRIQERLRDILSQWNHLLDLVSLRKSRLDAAVRYHQLFADADDIDNWMLDTLRLVSSEDTGVDEAQVQSLLKKHKDVTDELKNYANVIQQLKQQAQELSPEDANSGEVRERLAAIDARHGELAELARLRKQRLLDALSLFKLLAEADAADQWIAEKHRMLDTMLPPSDIDDVEIMKHRYDGFDKEMNANASRVAVVNQLARQLVHVEHPQAPRIQDRQHQLNHAWSALREKAEAKKDELKSAQGVQTFHIECRETVSWIEDKKRILQQTDDLEMDLNGVMTLQRRLSGMERDLAAIQARISSLESEASAIENEHPEEAQLIRERVEQITQNWEQLTHMLKERDSKLEEAGDLHRFLRSVDHFQAWLTKTQTDVASEDIPSSLPEAEKLLSQHQTIKEEIDNYKDEYAKMMEYGEKITAEPSTQDDPQYMFLRERLKALREGWAELQQMWENRQQLLAQSLELQLLQRDARQAEVLLAHQEHRLAKTEPPTNLEQAENIIKEHEAFLTTMEANDDKINSVVQFANRLVEERHFDADKIQRKAENIEARRNANREKALNLMEKLQDQLQLHQFLQDCDELGEWVQEKNVTAQDDTYRSAKTIHSKWTRHQAFEAEIAANKERLFAVQNAAEELMKQKPEFVEVISPKMHELQDQFENLQTTTKEKGERLFDANREVLLHQTCDDIDSWMNELEKQIQSTDTGSDLASVNILMQKQQMIETQMAVKAKQVTELETQAEYLQKTVPDKMEEIKEKKKTVEHRFEQLKAPLLERQRHLAKKKEAFQFRRDVEDEKLWVQEKMPLATSTDYGNSLFNVQMLQKKTQSLKTETDNHEPRILTVISNGQKLIDEGHESSPEFKQLIDELTARWRELKDAIEERKNNLSQWEKAHQYLFDANEAEAWMSEQELYMMVEDRGKDEISAQNLMKKHEILEQAVDDYAQTIRQLGETVRQLTAEEHPLSEQISVKQSQVDKLYAGLKDLAGERRAKLDEALRLFQLSREVDDLEQWITERELVASSQELGQDYDHVTLLWERFKEFARETQSVGSERVGTAERIADQMIAMGHSDNATIAQWKEGLKETWQDLLELIETRTQMLAASRELHKYFHDCKDTLQRVNERARGVSDELGRDAISVSALQRKHHNFMQDLTTLQQQVESIKSECSRLGASYAGEKAAEITRREREVADAWAALQAACRARRAKLEDADRLYRFLSQVRTLTLWMDDVVRQMNTGEKPRDVSGVELLMNNHQSLKAEIDTREDNFSACISLGKELLARQHYASADIKEKLLQLTNQRNALLRRWEERWENLQLILEVYQFARDAAVAEAWLIAQEPYLMSQELGHTIDEVESLIKKHEAFEKSAAAQEDRFSALQRLTTFEVKEMKRRQEAAEAAEREKREREAAEAAAAAAAAQDALDRADTPHEQPAANEASSVPNPAALAAPADPAAPAPPSPQQSTSKPTPVKPARLSKGASPASEDEGVEGTLVRKHEWESAAKRASNRSWDKLYVVAKDGRMSFYKDQKSYKSSPEQYWRGESPLDLNGAVVEVAANYTKKKHVFRLRLSSGAEFLLQAHDEAEMSWWLESLRARTQAPAARSHTLPAPANAEPKRRSFFTLKKN
Summary
Similarity
Belongs to the spectrin family.
Uniprot
A0A3S2NX91
A0A1E1W2D8
A0A194PGF0
A0A212F4U7
A0A2J7RAX6
A0A2J7RAX8
+ More
A0A2J7RAX9 A0A2J7RAY2 A0A2J7RAY1 A0A2J7RAX7 A0A2J7RAX1 A0A2J7RAX5 A0A067R2J7 A0A195EU35 E2ATM6 A0A158NGS2 A0A195BVX4 A0A158NGT0 A0A3L8DF89 A0A195DQE2 A0A026W2W3 A0A088ANZ7 E2C952 A0A232FMD3 A0A151X1T6 K7INW9 A0A1B6D863 A0A1B6CXW7 A0A0C9RDG7 A0A1B6CFD1 A0A1S3CZ92 B0WDS4 A0A1W4XCQ2 A0A154PNQ0 A0A1W4XCY0 A0A1W4XBL2 A0A1W4X295 A0A1Q3FIA9 A0A1Q3FIC1 W4VRI7 A0A1W4XCX5 A0A1Q3FIC3 A0A1Q3FI06 A0A2M3ZZA3 A0A2M3ZZ69 A0A069DY92 A0A224XH60 A0A310S8H6 Q178J6 A0A0P4VUS4 A0A0V0G3D5 A0A2M3ZZC5 W5JGW6 A0A023F5N6 A0A182FBX2 A0A2M3YXM3 A0A2M4B854 A0A2M4CM31 A0A2M4B842 E9IDX9 A0A1Q3FID4 A0A182PJD3 A0A1L8E5X8 A0A084VUR5 A0A1S4H577 Q7PSH4 A0A182WYC9 A0A182VAS0 A0A240PNM1 A0A2Y9D237 A0A182UC36 A0A182I5F7 A0A182INQ5 A0A182YKK4 A0A182K1X6 D2A0Z1 A0A182NCA0 A0A182WFY9 A0A1Y1KQM2 A0A182RW25 A0A182QW74 A0A2M4CG48 A0A182LU33 A0A0A9Z5K7 A0A0K8SG30 A0A0K8SEL8 A0A0A9Z7Y4 A0A0A9Z5L1 A0A0K8SEL9 A0A034W127 A0A0K8USQ3 A0A0L0CE19 T1HWC3 A0A0A1WFT8 E0VLF6 W8ANP9 A0A0T6B5L4 A0A1I8N998 A0A1I8N9A3 W8BL95
A0A2J7RAX9 A0A2J7RAY2 A0A2J7RAY1 A0A2J7RAX7 A0A2J7RAX1 A0A2J7RAX5 A0A067R2J7 A0A195EU35 E2ATM6 A0A158NGS2 A0A195BVX4 A0A158NGT0 A0A3L8DF89 A0A195DQE2 A0A026W2W3 A0A088ANZ7 E2C952 A0A232FMD3 A0A151X1T6 K7INW9 A0A1B6D863 A0A1B6CXW7 A0A0C9RDG7 A0A1B6CFD1 A0A1S3CZ92 B0WDS4 A0A1W4XCQ2 A0A154PNQ0 A0A1W4XCY0 A0A1W4XBL2 A0A1W4X295 A0A1Q3FIA9 A0A1Q3FIC1 W4VRI7 A0A1W4XCX5 A0A1Q3FIC3 A0A1Q3FI06 A0A2M3ZZA3 A0A2M3ZZ69 A0A069DY92 A0A224XH60 A0A310S8H6 Q178J6 A0A0P4VUS4 A0A0V0G3D5 A0A2M3ZZC5 W5JGW6 A0A023F5N6 A0A182FBX2 A0A2M3YXM3 A0A2M4B854 A0A2M4CM31 A0A2M4B842 E9IDX9 A0A1Q3FID4 A0A182PJD3 A0A1L8E5X8 A0A084VUR5 A0A1S4H577 Q7PSH4 A0A182WYC9 A0A182VAS0 A0A240PNM1 A0A2Y9D237 A0A182UC36 A0A182I5F7 A0A182INQ5 A0A182YKK4 A0A182K1X6 D2A0Z1 A0A182NCA0 A0A182WFY9 A0A1Y1KQM2 A0A182RW25 A0A182QW74 A0A2M4CG48 A0A182LU33 A0A0A9Z5K7 A0A0K8SG30 A0A0K8SEL8 A0A0A9Z7Y4 A0A0A9Z5L1 A0A0K8SEL9 A0A034W127 A0A0K8USQ3 A0A0L0CE19 T1HWC3 A0A0A1WFT8 E0VLF6 W8ANP9 A0A0T6B5L4 A0A1I8N998 A0A1I8N9A3 W8BL95
Pubmed
EMBL
RSAL01000035
RVE51333.1
GDQN01009940
GDQN01005007
JAT81114.1
JAT86047.1
+ More
KQ459604 KPI92466.1 AGBW02010317 OWR48743.1 NEVH01006564 PNF37986.1 PNF37985.1 PNF37984.1 PNF37982.1 PNF37988.1 PNF37981.1 PNF37983.1 PNF37989.1 KK852804 KDR16227.1 KQ981979 KYN31389.1 GL442629 EFN63222.1 ADTU01001618 KQ976398 KYM92779.1 QOIP01000009 RLU19125.1 KQ980612 KYN15088.1 KK107519 EZA49374.1 GL453780 EFN75525.1 NNAY01000039 OXU31669.1 KQ982585 KYQ54373.1 GEDC01015420 GEDC01011368 JAS21878.1 JAS25930.1 GEDC01019912 GEDC01018981 JAS17386.1 JAS18317.1 GBYB01011167 JAG80934.1 GEDC01025136 JAS12162.1 DS231900 EDS44827.1 KQ435007 KZC13483.1 GFDL01007749 JAV27296.1 GFDL01007748 JAV27297.1 GANO01002441 JAB57430.1 GFDL01007756 JAV27289.1 GFDL01007855 JAV27190.1 GGFK01000575 MBW33896.1 GGFK01000535 MBW33856.1 GBGD01000024 JAC88865.1 GFTR01008963 JAW07463.1 KQ770827 OAD52591.1 CH477362 EAT42646.1 GDKW01000285 JAI56310.1 GECL01003526 JAP02598.1 GGFK01000528 MBW33849.1 ADMH02001220 ETN63607.1 GBBI01002234 JAC16478.1 GGFM01000253 MBW21004.1 GGFJ01000066 MBW49207.1 GGFL01002161 MBW66339.1 GGFJ01000065 MBW49206.1 GL762556 EFZ21222.1 GFDL01007736 JAV27309.1 GFDF01000139 JAV13945.1 ATLV01016991 KE525139 KFB41709.1 AAAB01008823 EAA05634.3 APCN01003435 KQ971338 EFA01607.2 GEZM01081251 JAV61736.1 AXCN02001762 GGFL01000142 MBW64320.1 AXCM01008530 GBHO01003052 GBRD01014085 JAG40552.1 JAG51741.1 GBRD01014086 GDHC01017241 JAG51740.1 JAQ01388.1 GBRD01014084 JAG51742.1 GBHO01003045 JAG40559.1 GBHO01003047 JAG40557.1 GBRD01014083 JAG51743.1 GAKP01011465 GAKP01011464 JAC47488.1 GDHF01030988 GDHF01022939 GDHF01022145 GDHF01014185 GDHF01009959 GDHF01000114 JAI21326.1 JAI29375.1 JAI30169.1 JAI38129.1 JAI42355.1 JAI52200.1 JRES01000510 KNC30495.1 ACPB03018627 GBXI01017246 GBXI01016962 JAC97045.1 JAC97329.1 DS235271 EEB14212.1 GAMC01016135 JAB90420.1 LJIG01009657 KRT82642.1 GAMC01016136 JAB90419.1
KQ459604 KPI92466.1 AGBW02010317 OWR48743.1 NEVH01006564 PNF37986.1 PNF37985.1 PNF37984.1 PNF37982.1 PNF37988.1 PNF37981.1 PNF37983.1 PNF37989.1 KK852804 KDR16227.1 KQ981979 KYN31389.1 GL442629 EFN63222.1 ADTU01001618 KQ976398 KYM92779.1 QOIP01000009 RLU19125.1 KQ980612 KYN15088.1 KK107519 EZA49374.1 GL453780 EFN75525.1 NNAY01000039 OXU31669.1 KQ982585 KYQ54373.1 GEDC01015420 GEDC01011368 JAS21878.1 JAS25930.1 GEDC01019912 GEDC01018981 JAS17386.1 JAS18317.1 GBYB01011167 JAG80934.1 GEDC01025136 JAS12162.1 DS231900 EDS44827.1 KQ435007 KZC13483.1 GFDL01007749 JAV27296.1 GFDL01007748 JAV27297.1 GANO01002441 JAB57430.1 GFDL01007756 JAV27289.1 GFDL01007855 JAV27190.1 GGFK01000575 MBW33896.1 GGFK01000535 MBW33856.1 GBGD01000024 JAC88865.1 GFTR01008963 JAW07463.1 KQ770827 OAD52591.1 CH477362 EAT42646.1 GDKW01000285 JAI56310.1 GECL01003526 JAP02598.1 GGFK01000528 MBW33849.1 ADMH02001220 ETN63607.1 GBBI01002234 JAC16478.1 GGFM01000253 MBW21004.1 GGFJ01000066 MBW49207.1 GGFL01002161 MBW66339.1 GGFJ01000065 MBW49206.1 GL762556 EFZ21222.1 GFDL01007736 JAV27309.1 GFDF01000139 JAV13945.1 ATLV01016991 KE525139 KFB41709.1 AAAB01008823 EAA05634.3 APCN01003435 KQ971338 EFA01607.2 GEZM01081251 JAV61736.1 AXCN02001762 GGFL01000142 MBW64320.1 AXCM01008530 GBHO01003052 GBRD01014085 JAG40552.1 JAG51741.1 GBRD01014086 GDHC01017241 JAG51740.1 JAQ01388.1 GBRD01014084 JAG51742.1 GBHO01003045 JAG40559.1 GBHO01003047 JAG40557.1 GBRD01014083 JAG51743.1 GAKP01011465 GAKP01011464 JAC47488.1 GDHF01030988 GDHF01022939 GDHF01022145 GDHF01014185 GDHF01009959 GDHF01000114 JAI21326.1 JAI29375.1 JAI30169.1 JAI38129.1 JAI42355.1 JAI52200.1 JRES01000510 KNC30495.1 ACPB03018627 GBXI01017246 GBXI01016962 JAC97045.1 JAC97329.1 DS235271 EEB14212.1 GAMC01016135 JAB90420.1 LJIG01009657 KRT82642.1 GAMC01016136 JAB90419.1
Proteomes
UP000283053
UP000053268
UP000007151
UP000235965
UP000027135
UP000078541
+ More
UP000000311 UP000005205 UP000078540 UP000279307 UP000078492 UP000053097 UP000005203 UP000008237 UP000215335 UP000075809 UP000002358 UP000079169 UP000002320 UP000192223 UP000076502 UP000008820 UP000000673 UP000069272 UP000075885 UP000030765 UP000007062 UP000076407 UP000075903 UP000075880 UP000075902 UP000075840 UP000076408 UP000075881 UP000007266 UP000075884 UP000075920 UP000075900 UP000075886 UP000075883 UP000037069 UP000015103 UP000009046 UP000095301
UP000000311 UP000005205 UP000078540 UP000279307 UP000078492 UP000053097 UP000005203 UP000008237 UP000215335 UP000075809 UP000002358 UP000079169 UP000002320 UP000192223 UP000076502 UP000008820 UP000000673 UP000069272 UP000075885 UP000030765 UP000007062 UP000076407 UP000075903 UP000075880 UP000075902 UP000075840 UP000076408 UP000075881 UP000007266 UP000075884 UP000075920 UP000075900 UP000075886 UP000075883 UP000037069 UP000015103 UP000009046 UP000095301
Pfam
Interpro
IPR001849
PH_domain
+ More
IPR001589 Actinin_actin-bd_CS
IPR011993 PH-like_dom_sf
IPR041681 PH_9
IPR002017 Spectrin_repeat
IPR016343 Spectrin_bsu
IPR001715 CH-domain
IPR036872 CH_dom_sf
IPR018159 Spectrin/alpha-actinin
IPR001605 PH_dom-spectrin-type
IPR006581 VPS10
IPR011042 6-blade_b-propeller_TolB-like
IPR000742 EGF-like_dom
IPR023415 LDLR_class-A_CS
IPR031777 Sortilin_C
IPR000033 LDLR_classB_rpt
IPR031778 Sortilin_N
IPR002172 LDrepeatLR_classA_rpt
IPR036116 FN3_sf
IPR036055 LDL_receptor-like_sf
IPR003961 FN3_dom
IPR013783 Ig-like_fold
IPR001589 Actinin_actin-bd_CS
IPR011993 PH-like_dom_sf
IPR041681 PH_9
IPR002017 Spectrin_repeat
IPR016343 Spectrin_bsu
IPR001715 CH-domain
IPR036872 CH_dom_sf
IPR018159 Spectrin/alpha-actinin
IPR001605 PH_dom-spectrin-type
IPR006581 VPS10
IPR011042 6-blade_b-propeller_TolB-like
IPR000742 EGF-like_dom
IPR023415 LDLR_class-A_CS
IPR031777 Sortilin_C
IPR000033 LDLR_classB_rpt
IPR031778 Sortilin_N
IPR002172 LDrepeatLR_classA_rpt
IPR036116 FN3_sf
IPR036055 LDL_receptor-like_sf
IPR003961 FN3_dom
IPR013783 Ig-like_fold
ProteinModelPortal
A0A3S2NX91
A0A1E1W2D8
A0A194PGF0
A0A212F4U7
A0A2J7RAX6
A0A2J7RAX8
+ More
A0A2J7RAX9 A0A2J7RAY2 A0A2J7RAY1 A0A2J7RAX7 A0A2J7RAX1 A0A2J7RAX5 A0A067R2J7 A0A195EU35 E2ATM6 A0A158NGS2 A0A195BVX4 A0A158NGT0 A0A3L8DF89 A0A195DQE2 A0A026W2W3 A0A088ANZ7 E2C952 A0A232FMD3 A0A151X1T6 K7INW9 A0A1B6D863 A0A1B6CXW7 A0A0C9RDG7 A0A1B6CFD1 A0A1S3CZ92 B0WDS4 A0A1W4XCQ2 A0A154PNQ0 A0A1W4XCY0 A0A1W4XBL2 A0A1W4X295 A0A1Q3FIA9 A0A1Q3FIC1 W4VRI7 A0A1W4XCX5 A0A1Q3FIC3 A0A1Q3FI06 A0A2M3ZZA3 A0A2M3ZZ69 A0A069DY92 A0A224XH60 A0A310S8H6 Q178J6 A0A0P4VUS4 A0A0V0G3D5 A0A2M3ZZC5 W5JGW6 A0A023F5N6 A0A182FBX2 A0A2M3YXM3 A0A2M4B854 A0A2M4CM31 A0A2M4B842 E9IDX9 A0A1Q3FID4 A0A182PJD3 A0A1L8E5X8 A0A084VUR5 A0A1S4H577 Q7PSH4 A0A182WYC9 A0A182VAS0 A0A240PNM1 A0A2Y9D237 A0A182UC36 A0A182I5F7 A0A182INQ5 A0A182YKK4 A0A182K1X6 D2A0Z1 A0A182NCA0 A0A182WFY9 A0A1Y1KQM2 A0A182RW25 A0A182QW74 A0A2M4CG48 A0A182LU33 A0A0A9Z5K7 A0A0K8SG30 A0A0K8SEL8 A0A0A9Z7Y4 A0A0A9Z5L1 A0A0K8SEL9 A0A034W127 A0A0K8USQ3 A0A0L0CE19 T1HWC3 A0A0A1WFT8 E0VLF6 W8ANP9 A0A0T6B5L4 A0A1I8N998 A0A1I8N9A3 W8BL95
A0A2J7RAX9 A0A2J7RAY2 A0A2J7RAY1 A0A2J7RAX7 A0A2J7RAX1 A0A2J7RAX5 A0A067R2J7 A0A195EU35 E2ATM6 A0A158NGS2 A0A195BVX4 A0A158NGT0 A0A3L8DF89 A0A195DQE2 A0A026W2W3 A0A088ANZ7 E2C952 A0A232FMD3 A0A151X1T6 K7INW9 A0A1B6D863 A0A1B6CXW7 A0A0C9RDG7 A0A1B6CFD1 A0A1S3CZ92 B0WDS4 A0A1W4XCQ2 A0A154PNQ0 A0A1W4XCY0 A0A1W4XBL2 A0A1W4X295 A0A1Q3FIA9 A0A1Q3FIC1 W4VRI7 A0A1W4XCX5 A0A1Q3FIC3 A0A1Q3FI06 A0A2M3ZZA3 A0A2M3ZZ69 A0A069DY92 A0A224XH60 A0A310S8H6 Q178J6 A0A0P4VUS4 A0A0V0G3D5 A0A2M3ZZC5 W5JGW6 A0A023F5N6 A0A182FBX2 A0A2M3YXM3 A0A2M4B854 A0A2M4CM31 A0A2M4B842 E9IDX9 A0A1Q3FID4 A0A182PJD3 A0A1L8E5X8 A0A084VUR5 A0A1S4H577 Q7PSH4 A0A182WYC9 A0A182VAS0 A0A240PNM1 A0A2Y9D237 A0A182UC36 A0A182I5F7 A0A182INQ5 A0A182YKK4 A0A182K1X6 D2A0Z1 A0A182NCA0 A0A182WFY9 A0A1Y1KQM2 A0A182RW25 A0A182QW74 A0A2M4CG48 A0A182LU33 A0A0A9Z5K7 A0A0K8SG30 A0A0K8SEL8 A0A0A9Z7Y4 A0A0A9Z5L1 A0A0K8SEL9 A0A034W127 A0A0K8USQ3 A0A0L0CE19 T1HWC3 A0A0A1WFT8 E0VLF6 W8ANP9 A0A0T6B5L4 A0A1I8N998 A0A1I8N9A3 W8BL95
PDB
6ANU
E-value=2.02782e-111,
Score=1036
Ontologies
GO
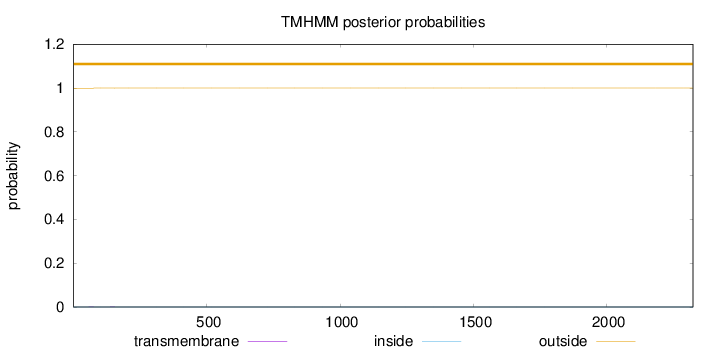
Topology
Subcellular location
Golgi apparatus
Length:
2323
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00845999999999998
Exp number, first 60 AAs:
0.00038
Total prob of N-in:
0.00046
outside
1 - 2323
Population Genetic Test Statistics
Pi
285.605821
Theta
178.775471
Tajima's D
1.868076
CLR
0.143998
CSRT
0.864606769661517
Interpretation
Uncertain
