Gene
KWMTBOMO11042
Annotation
chymotrypsin-like_serine_protease_precursor_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.003 PlasmaMembrane Reliability : 2.653
Sequence
CDS
ATGAAAACTGCTGTACTCTTCGGTATCTTCTTCGTGGGCTACACATCAGCCTATGTGCCGGTGGAGACCTTCTACCACGAGAATGTGGGAATACCCCTAGCTAAAAGTATTAGGGCTGCGGAGACTGCAAAGCTTGACTCAAGTGTCCAGCCAGACAATGCTGCAAGGATAGTTGGTGGGGCCATTTCGCCATCAAACGCACATCCTTATTTGGCTGGTCTTCTCATCACCTTCATAAACGCCGTCGGTACTTCGGCGTGTGGATCATCGCTTCTTTCAGCTAACAGGCTAGTCACCGCCGCCCACTGCTGGTTTGATGGCCGCTTCCAAGCTAACCAGTTCGTTGTGGTTCTCGGCTCAAACACGCTGTTCCACGGTGGTGTTCGTGTCACTACTCGTCAAGTTTTTGTACATCCTCAATGGAATCCCACCCTTTTGAACAACGATGTGGCAATGATTTACCTACCTCACAGGGTTACTCTGAACAACAACATCAAGCCCATCGCTCTGCCTAACACCGCCGATTTGAACAACCTATTTGTTGGCCAATGGGCGGTCGCTGCTGGATATGGATTAACTAGTGACGCGCAAACTGGTATCTCGGTAAATCAGGTGATGAGCCAAGTGAACCTGCAAGTGATCACAGTGCAACAGTGCATGGCCGTTTTCGGTAGCAACTTCGTGAGGAACTCAAACATCTGCACGAACGGAGCTGGCGGTGTCGGTATTTGCCGCGGAGACTCCGGCGGACCACTTTTGTTGAACCGTAATGGTGTACTAACCTTAATCGGTATCAGTTCTTTCGTAGCCCAGAACAGATGCCAGGACGGATTCCCATCTGCCTTCGCCAGAGTTACAAGTTTCAACAATTTCATCCGCCAGCATTTATAA
Protein
MKTAVLFGIFFVGYTSAYVPVETFYHENVGIPLAKSIRAAETAKLDSSVQPDNAARIVGGAISPSNAHPYLAGLLITFINAVGTSACGSSLLSANRLVTAAHCWFDGRFQANQFVVVLGSNTLFHGGVRVTTRQVFVHPQWNPTLLNNDVAMIYLPHRVTLNNNIKPIALPNTADLNNLFVGQWAVAAGYGLTSDAQTGISVNQVMSQVNLQVITVQQCMAVFGSNFVRNSNICTNGAGGVGICRGDSGGPLLLNRNGVLTLIGISSFVAQNRCQDGFPSAFARVTSFNNFIRQHL
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
Q1HPW8
A0A3S2NMC5
A0A1B1LTR9
A0A3S2LDG1
I6TMX9
I6SJ44
+ More
A0A0L7LI04 A0A2H1X112 A0A1E1W2N0 A0A1E1WL73 A0A194PHE1 A0A1E1WB13 A0A0N0PBU1 A0A0M4MQT7 A0A1E1WTE4 A0A2A4JK09 A0A2A4JJ19 A0A1E1WDT0 A0A089Q9M9 A0A2A4JJV5 A0A2H1X2D3 A0A212F096 A0A2A4JJQ3 C9W8J3 E0W9N7 C9W8G2 C9W8H9 A0A2A4JIX9 A0A2A4JKX6 C9W8G9 A0A0M5KXW7 A0A194PHC5 E7D016 A0A194R1N4 A0A1E1WJ19 J7FDD9 A0A089QAL5 A0A0K8SR13 A0A089QDB8 A0A2W1BWV2 C9W8J4 A0A2H1V6W6 A0A2A4K0B4 Q2F617 A0PGD5 A0A194Q0R8 J7FCZ4 A0A2A4JFK9 J7FDV8 A0A0N1PGY7 O18438 C9W8F5 A0A2H1V7A9 Q9NGY4 A0A2H1V6P6 A0A089RSY0 H9JG67 O18444 C7FF06 A0A2H1WCW9 A0A2W1BAG4 O18450 A0A2A4JFM8 A0PGD4 Q9NH07 E7D003 C9W8G3 A0A2A4K202 A0A2H1VX12 C9W8F6 O18445 A0A0K8SS51 A0A142I706 E0W9N4 I6SJ48 A0A2H1VAW7 C9W8G8 A0A2A4K1Q8 B4X989 H9JM32 A0PGD7 D7RZZ7 C9W8G7 A0A1B0RHP3 E7D005 Q86M89 B4X991 A0A2A4K1C2 A0A2A4K1R6 A0A0L7L6E9 A0A0L7LB31 J7FDV3 A0A2A4JSJ6 D7RZZ8 A0A089QAK4 A0A2A4K228 A0A2A4K0D3 A0A0X9HJY6 O18443 D7RZZ6
A0A0L7LI04 A0A2H1X112 A0A1E1W2N0 A0A1E1WL73 A0A194PHE1 A0A1E1WB13 A0A0N0PBU1 A0A0M4MQT7 A0A1E1WTE4 A0A2A4JK09 A0A2A4JJ19 A0A1E1WDT0 A0A089Q9M9 A0A2A4JJV5 A0A2H1X2D3 A0A212F096 A0A2A4JJQ3 C9W8J3 E0W9N7 C9W8G2 C9W8H9 A0A2A4JIX9 A0A2A4JKX6 C9W8G9 A0A0M5KXW7 A0A194PHC5 E7D016 A0A194R1N4 A0A1E1WJ19 J7FDD9 A0A089QAL5 A0A0K8SR13 A0A089QDB8 A0A2W1BWV2 C9W8J4 A0A2H1V6W6 A0A2A4K0B4 Q2F617 A0PGD5 A0A194Q0R8 J7FCZ4 A0A2A4JFK9 J7FDV8 A0A0N1PGY7 O18438 C9W8F5 A0A2H1V7A9 Q9NGY4 A0A2H1V6P6 A0A089RSY0 H9JG67 O18444 C7FF06 A0A2H1WCW9 A0A2W1BAG4 O18450 A0A2A4JFM8 A0PGD4 Q9NH07 E7D003 C9W8G3 A0A2A4K202 A0A2H1VX12 C9W8F6 O18445 A0A0K8SS51 A0A142I706 E0W9N4 I6SJ48 A0A2H1VAW7 C9W8G8 A0A2A4K1Q8 B4X989 H9JM32 A0PGD7 D7RZZ7 C9W8G7 A0A1B0RHP3 E7D005 Q86M89 B4X991 A0A2A4K1C2 A0A2A4K1R6 A0A0L7L6E9 A0A0L7LB31 J7FDV3 A0A2A4JSJ6 D7RZZ8 A0A089QAK4 A0A2A4K228 A0A2A4K0D3 A0A0X9HJY6 O18443 D7RZZ6
Pubmed
EMBL
DQ443284
ABF51373.1
RSAL01000035
RVE51335.1
KX380993
ANS56509.1
+ More
RVE51334.1 JQ904138 AFM77768.1 JQ904137 AFM77767.1 JTDY01001059 KOB74999.1 ODYU01012588 SOQ59001.1 GDQN01009850 JAT81204.1 GDQN01003329 JAT87725.1 KQ459604 KPI92468.1 GDQN01006881 JAT84173.1 KQ460813 KPJ12139.1 KR024678 ALE15220.1 GDQN01000810 JAT90244.1 NWSH01001222 PCG72058.1 PCG72057.1 GDQN01005957 JAT85097.1 KM083783 AIR09764.1 PCG72056.1 ODYU01012894 SOQ59427.1 AGBW02011133 OWR47160.1 PCG72059.1 FJ205442 ACR16006.2 FJ205439 ADM35108.1 FJ205407 ACR15975.2 FJ205425 ACR15992.2 PCG72055.1 PCG72060.1 FJ205414 ACR15982.2 KR024677 ALE15219.1 KQ459603 KPI92811.1 HM990186 ADT80834.1 KQ460883 KPJ11150.1 GDQN01004055 JAT86999.1 JN252038 AFM28251.1 KM083794 AIR09775.1 GBRD01010002 JAG55822.1 KM083792 AIR09773.1 KZ149911 PZC78104.1 FJ205444 ACR16007.1 ODYU01000971 SOQ36526.1 NWSH01000291 PCG77701.1 DQ311255 ABD36200.1 AY618890 AAV33653.1 KQ459581 KPI99177.1 JN252036 AFM28249.1 NWSH01001552 PCG70875.1 JN252045 AFM28258.1 KQ461033 KPJ09997.1 Y12273 CAA72952.1 FJ205399 ACR15968.2 ODYU01001034 SOQ36689.1 AF237417 EF531621 AAF43709.1 ABR88232.1 SOQ36525.1 KM083781 AIR09762.1 BABH01002097 Y12280 CAA72959.1 GQ354838 ACU00133.1 ODYU01007826 SOQ50918.1 KZ150191 PZC72392.1 Y12287 CAA72966.1 PCG70877.1 AY618889 AAV33652.1 AF233732 AAF71519.1 HM990173 ADT80821.1 FJ205408 ACR15976.1 PCG77700.1 ODYU01004792 SOQ45042.1 FJ205400 ACR15969.2 Y12281 CAA72960.1 GBRD01009676 JAG56148.1 KT907053 AMR44225.1 FJ205401 ADM35105.1 JQ904142 AFM77772.1 ODYU01001570 SOQ37967.1 FJ205413 ACR15981.1 PCG77693.1 EF531625 ABR88236.1 BABH01005812 AY618892 AAV33655.1 HM209420 ADI32881.1 FJ205412 ACR15980.1 KM360186 AKH49602.1 HM990175 ADT80823.1 AY251276 AAO75039.1 EF531627 ABR88238.1 PCG77694.1 PCG77703.1 JTDY01002618 KOB71083.1 JTDY01001894 KOB72615.1 JN252035 AFM28248.1 NWSH01000651 PCG75015.1 HM209421 ADI32882.1 KM083784 AIR09765.1 PCG77692.1 PCG77697.1 KP690081 ALO61083.1 SOQ36527.1 Y12279 CAA72958.1 HM209419 ADI32880.1
RVE51334.1 JQ904138 AFM77768.1 JQ904137 AFM77767.1 JTDY01001059 KOB74999.1 ODYU01012588 SOQ59001.1 GDQN01009850 JAT81204.1 GDQN01003329 JAT87725.1 KQ459604 KPI92468.1 GDQN01006881 JAT84173.1 KQ460813 KPJ12139.1 KR024678 ALE15220.1 GDQN01000810 JAT90244.1 NWSH01001222 PCG72058.1 PCG72057.1 GDQN01005957 JAT85097.1 KM083783 AIR09764.1 PCG72056.1 ODYU01012894 SOQ59427.1 AGBW02011133 OWR47160.1 PCG72059.1 FJ205442 ACR16006.2 FJ205439 ADM35108.1 FJ205407 ACR15975.2 FJ205425 ACR15992.2 PCG72055.1 PCG72060.1 FJ205414 ACR15982.2 KR024677 ALE15219.1 KQ459603 KPI92811.1 HM990186 ADT80834.1 KQ460883 KPJ11150.1 GDQN01004055 JAT86999.1 JN252038 AFM28251.1 KM083794 AIR09775.1 GBRD01010002 JAG55822.1 KM083792 AIR09773.1 KZ149911 PZC78104.1 FJ205444 ACR16007.1 ODYU01000971 SOQ36526.1 NWSH01000291 PCG77701.1 DQ311255 ABD36200.1 AY618890 AAV33653.1 KQ459581 KPI99177.1 JN252036 AFM28249.1 NWSH01001552 PCG70875.1 JN252045 AFM28258.1 KQ461033 KPJ09997.1 Y12273 CAA72952.1 FJ205399 ACR15968.2 ODYU01001034 SOQ36689.1 AF237417 EF531621 AAF43709.1 ABR88232.1 SOQ36525.1 KM083781 AIR09762.1 BABH01002097 Y12280 CAA72959.1 GQ354838 ACU00133.1 ODYU01007826 SOQ50918.1 KZ150191 PZC72392.1 Y12287 CAA72966.1 PCG70877.1 AY618889 AAV33652.1 AF233732 AAF71519.1 HM990173 ADT80821.1 FJ205408 ACR15976.1 PCG77700.1 ODYU01004792 SOQ45042.1 FJ205400 ACR15969.2 Y12281 CAA72960.1 GBRD01009676 JAG56148.1 KT907053 AMR44225.1 FJ205401 ADM35105.1 JQ904142 AFM77772.1 ODYU01001570 SOQ37967.1 FJ205413 ACR15981.1 PCG77693.1 EF531625 ABR88236.1 BABH01005812 AY618892 AAV33655.1 HM209420 ADI32881.1 FJ205412 ACR15980.1 KM360186 AKH49602.1 HM990175 ADT80823.1 AY251276 AAO75039.1 EF531627 ABR88238.1 PCG77694.1 PCG77703.1 JTDY01002618 KOB71083.1 JTDY01001894 KOB72615.1 JN252035 AFM28248.1 NWSH01000651 PCG75015.1 HM209421 ADI32882.1 KM083784 AIR09765.1 PCG77692.1 PCG77697.1 KP690081 ALO61083.1 SOQ36527.1 Y12279 CAA72958.1 HM209419 ADI32880.1
Proteomes
Pfam
PF00089 Trypsin
Interpro
CDD
ProteinModelPortal
Q1HPW8
A0A3S2NMC5
A0A1B1LTR9
A0A3S2LDG1
I6TMX9
I6SJ44
+ More
A0A0L7LI04 A0A2H1X112 A0A1E1W2N0 A0A1E1WL73 A0A194PHE1 A0A1E1WB13 A0A0N0PBU1 A0A0M4MQT7 A0A1E1WTE4 A0A2A4JK09 A0A2A4JJ19 A0A1E1WDT0 A0A089Q9M9 A0A2A4JJV5 A0A2H1X2D3 A0A212F096 A0A2A4JJQ3 C9W8J3 E0W9N7 C9W8G2 C9W8H9 A0A2A4JIX9 A0A2A4JKX6 C9W8G9 A0A0M5KXW7 A0A194PHC5 E7D016 A0A194R1N4 A0A1E1WJ19 J7FDD9 A0A089QAL5 A0A0K8SR13 A0A089QDB8 A0A2W1BWV2 C9W8J4 A0A2H1V6W6 A0A2A4K0B4 Q2F617 A0PGD5 A0A194Q0R8 J7FCZ4 A0A2A4JFK9 J7FDV8 A0A0N1PGY7 O18438 C9W8F5 A0A2H1V7A9 Q9NGY4 A0A2H1V6P6 A0A089RSY0 H9JG67 O18444 C7FF06 A0A2H1WCW9 A0A2W1BAG4 O18450 A0A2A4JFM8 A0PGD4 Q9NH07 E7D003 C9W8G3 A0A2A4K202 A0A2H1VX12 C9W8F6 O18445 A0A0K8SS51 A0A142I706 E0W9N4 I6SJ48 A0A2H1VAW7 C9W8G8 A0A2A4K1Q8 B4X989 H9JM32 A0PGD7 D7RZZ7 C9W8G7 A0A1B0RHP3 E7D005 Q86M89 B4X991 A0A2A4K1C2 A0A2A4K1R6 A0A0L7L6E9 A0A0L7LB31 J7FDV3 A0A2A4JSJ6 D7RZZ8 A0A089QAK4 A0A2A4K228 A0A2A4K0D3 A0A0X9HJY6 O18443 D7RZZ6
A0A0L7LI04 A0A2H1X112 A0A1E1W2N0 A0A1E1WL73 A0A194PHE1 A0A1E1WB13 A0A0N0PBU1 A0A0M4MQT7 A0A1E1WTE4 A0A2A4JK09 A0A2A4JJ19 A0A1E1WDT0 A0A089Q9M9 A0A2A4JJV5 A0A2H1X2D3 A0A212F096 A0A2A4JJQ3 C9W8J3 E0W9N7 C9W8G2 C9W8H9 A0A2A4JIX9 A0A2A4JKX6 C9W8G9 A0A0M5KXW7 A0A194PHC5 E7D016 A0A194R1N4 A0A1E1WJ19 J7FDD9 A0A089QAL5 A0A0K8SR13 A0A089QDB8 A0A2W1BWV2 C9W8J4 A0A2H1V6W6 A0A2A4K0B4 Q2F617 A0PGD5 A0A194Q0R8 J7FCZ4 A0A2A4JFK9 J7FDV8 A0A0N1PGY7 O18438 C9W8F5 A0A2H1V7A9 Q9NGY4 A0A2H1V6P6 A0A089RSY0 H9JG67 O18444 C7FF06 A0A2H1WCW9 A0A2W1BAG4 O18450 A0A2A4JFM8 A0PGD4 Q9NH07 E7D003 C9W8G3 A0A2A4K202 A0A2H1VX12 C9W8F6 O18445 A0A0K8SS51 A0A142I706 E0W9N4 I6SJ48 A0A2H1VAW7 C9W8G8 A0A2A4K1Q8 B4X989 H9JM32 A0PGD7 D7RZZ7 C9W8G7 A0A1B0RHP3 E7D005 Q86M89 B4X991 A0A2A4K1C2 A0A2A4K1R6 A0A0L7L6E9 A0A0L7LB31 J7FDV3 A0A2A4JSJ6 D7RZZ8 A0A089QAK4 A0A2A4K228 A0A2A4K0D3 A0A0X9HJY6 O18443 D7RZZ6
PDB
2HLC
E-value=4.96268e-30,
Score=325
Ontologies
GO
Topology
SignalP
Position: 1 - 17,
Likelihood: 0.986387
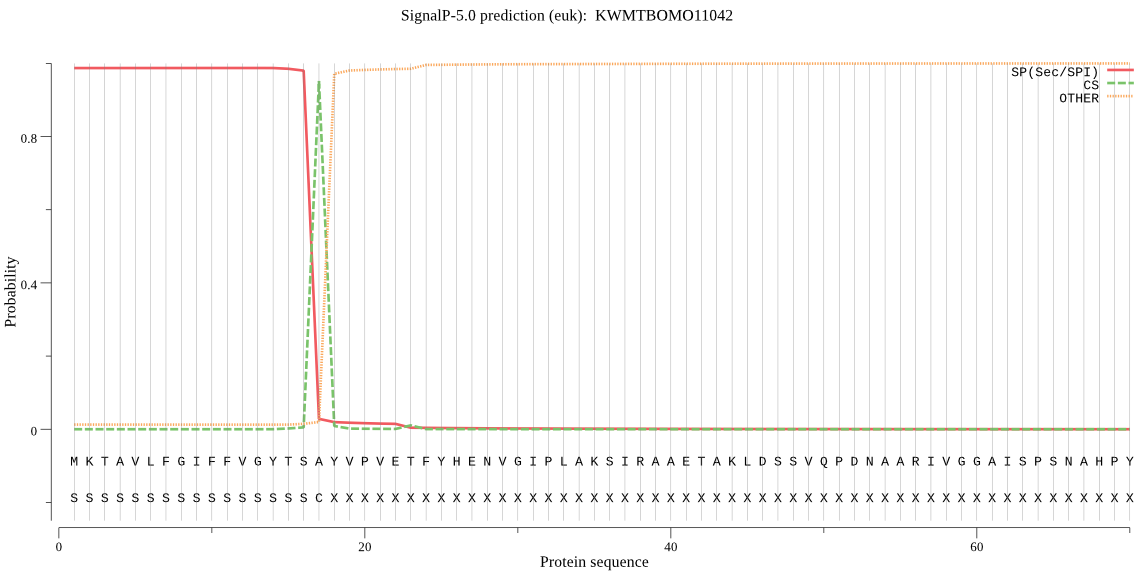
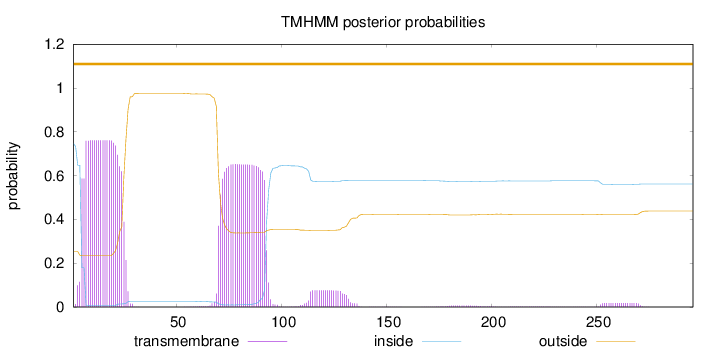
Length:
296
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
32.3146
Exp number, first 60 AAs:
15.4447
Total prob of N-in:
0.74651
POSSIBLE N-term signal
sequence
outside
1 - 296
Population Genetic Test Statistics
Pi
226.829263
Theta
183.298504
Tajima's D
0.782398
CLR
0.014609
CSRT
0.59662016899155
Interpretation
Uncertain
