Gene
KWMTBOMO11041
Pre Gene Modal
BGIBMGA008541
Annotation
hypothetical_protein_KGM_10985_[Danaus_plexippus]
Location in the cell
Nuclear Reliability : 2.606
Sequence
CDS
ATGTGGTGGACAATCACTGGCAACTTCGGGAACATCTTACCTATTGACTGGACCAAATCTTTCTCGAGGAAGATGCATATTCCTACATTAAATTTGAGTGATACAAAGAATTCGTTGACTCCGGATGACGAGATCCACAGCTCAGAAGACGAGGCTGTCGCCTCAGACTTGGACATGCACGCTCTTATCCTGGGCGGCCTTCATCAGGACCACGAGTGCCCTGTCAAGACTGCTGACGAAGTGATTCAAGAAATAGACGATATGATGCAAGACACTCCGTCGTCGGAGGGTGAATACATAGAGTACAATGAGGCACTCGAGAAAGGCAAGGAGGTGCTCAACTCCCCCCTCTATGAAGACAAGCTGCGCGCATTATCTTTAGCTCAATTAAACGAGATCTTCATGGAACTGGAGGTGTTGATCCGGGAATTCTCCGAGACCTTGATAGCGGAACTCGCTCTACGGGACGAGCTGGAGTACGAGAAGGAAATGAAAAACACTTTCATATCACTCCTGCTGGCCGTGCAGAACAAACGCAGGCAGCATTACGTCGAGAAGAAGAAGAGGCCCGGTGCGCCGCGCTCGCCGAACCTCACCAGCAACAGCTCCAAGCCGAAGTACTTGACCACCGTCATACCATTCCACTTGGACAACGGACCACCAGACAACCAGTCACTTCAAGTACTCATCAAAATACTGAAAGCTATAAACGAAGACAGCCCCACCGTGCCTACTCTACTCACAGACTACATCCTGAAGGTGTTGTGTCCAACATAA
Protein
MWWTITGNFGNILPIDWTKSFSRKMHIPTLNLSDTKNSLTPDDEIHSSEDEAVASDLDMHALILGGLHQDHECPVKTADEVIQEIDDMMQDTPSSEGEYIEYNEALEKGKEVLNSPLYEDKLRALSLAQLNEIFMELEVLIREFSETLIAELALRDELEYEKEMKNTFISLLLAVQNKRRQHYVEKKKRPGAPRSPNLTSNSSKPKYLTTVIPFHLDNGPPDNQSLQVLIKILKAINEDSPTVPTLLTDYILKVLCPT
Summary
Uniprot
A0A3S2TP34
S4P6I4
A0A2A4JKD1
A0A212F0D3
A0A0N1IP27
H9JG94
+ More
A0A194PIB7 A0A1Y1KAH8 A0A1E1WTM4 A0A139WJ97 A0A139WJ27 V5I8M4 A0A2J7PC04 U5ESI1 A0A1J1IHQ8 A0A336M0C6 Q172Q9 A0A182GUG2 A0A023ETZ5 A0A182SL00 A0A182LFT5 A0A182VC56 A0A182HU79 A0A182TXW6 Q7QD97 A0A182WRJ7 A0A182P5T5 A0A182RNR3 A0A182W064 A0A182K945 A0A182QRB0 A0A182N640 U4U321 A0A0K8TM88 A0A182LZF5 N6SYH4 A0A182Y4A7 A0A2M3Z305 A0A1B6HPW0 A0A182IYB7 A0A084VRB9 A0A2M3Z4U8 A0A2M4BRP9 A0A224XSN3 A0A1B6F6T4 W5J514 A0A2M4BPA6 A0A2M4BNE6 A0A067RIX0 A0A0P4VHV2 A0A023F4Z1 A0A2M4AES3 A0A1B6DQJ0 T1E205 A0A1Q3FXE4 A0A0A1XGK8 A0A034VCN7 A0A0K8UQR5 W8B648 A0A146L607 A0A0A9WPQ4 A0A0K8SPF7 A0A0M4F8V1 A0A158NH83 A0A182FRE8 A0A026WA38 T1HXL5 E0VM28 A0A2A3ECR3 V9IDS1 A0A088AHM0 A0A0L0C2P0 A0A1B0FCH7 A0A1A9Z6D1 A0A1A9USZ9 A0A1B0B611 A0A1I8PX97 T1PH91 A0A1I8MLI2 B4L1E3 B4JM48 B4M775 A0A0J7L9M2 A0A195B7P7 A0A0N0BIL8 E2AL25 M9PGS9 K7IXY2 A0A232EM24 A0A2S2N7N8 A0A1A9X5M8 A0A0C9RMJ6 Q29I15 A0A1L8DVX9 A0A1B0CDT6 B4Q1B3 A0A195FEM8 B4NBW0 B3P8W1
A0A194PIB7 A0A1Y1KAH8 A0A1E1WTM4 A0A139WJ97 A0A139WJ27 V5I8M4 A0A2J7PC04 U5ESI1 A0A1J1IHQ8 A0A336M0C6 Q172Q9 A0A182GUG2 A0A023ETZ5 A0A182SL00 A0A182LFT5 A0A182VC56 A0A182HU79 A0A182TXW6 Q7QD97 A0A182WRJ7 A0A182P5T5 A0A182RNR3 A0A182W064 A0A182K945 A0A182QRB0 A0A182N640 U4U321 A0A0K8TM88 A0A182LZF5 N6SYH4 A0A182Y4A7 A0A2M3Z305 A0A1B6HPW0 A0A182IYB7 A0A084VRB9 A0A2M3Z4U8 A0A2M4BRP9 A0A224XSN3 A0A1B6F6T4 W5J514 A0A2M4BPA6 A0A2M4BNE6 A0A067RIX0 A0A0P4VHV2 A0A023F4Z1 A0A2M4AES3 A0A1B6DQJ0 T1E205 A0A1Q3FXE4 A0A0A1XGK8 A0A034VCN7 A0A0K8UQR5 W8B648 A0A146L607 A0A0A9WPQ4 A0A0K8SPF7 A0A0M4F8V1 A0A158NH83 A0A182FRE8 A0A026WA38 T1HXL5 E0VM28 A0A2A3ECR3 V9IDS1 A0A088AHM0 A0A0L0C2P0 A0A1B0FCH7 A0A1A9Z6D1 A0A1A9USZ9 A0A1B0B611 A0A1I8PX97 T1PH91 A0A1I8MLI2 B4L1E3 B4JM48 B4M775 A0A0J7L9M2 A0A195B7P7 A0A0N0BIL8 E2AL25 M9PGS9 K7IXY2 A0A232EM24 A0A2S2N7N8 A0A1A9X5M8 A0A0C9RMJ6 Q29I15 A0A1L8DVX9 A0A1B0CDT6 B4Q1B3 A0A195FEM8 B4NBW0 B3P8W1
Pubmed
23622113
22118469
26354079
19121390
28004739
18362917
+ More
19820115 17510324 26483478 24945155 20966253 12364791 14747013 17210077 23537049 26369729 25244985 24438588 20920257 23761445 24845553 27129103 25474469 24330624 25830018 25348373 24495485 26823975 25401762 21347285 24508170 30249741 20566863 26108605 25315136 17994087 20798317 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 20075255 28648823 15632085 17550304
19820115 17510324 26483478 24945155 20966253 12364791 14747013 17210077 23537049 26369729 25244985 24438588 20920257 23761445 24845553 27129103 25474469 24330624 25830018 25348373 24495485 26823975 25401762 21347285 24508170 30249741 20566863 26108605 25315136 17994087 20798317 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 20075255 28648823 15632085 17550304
EMBL
RSAL01000035
RVE51337.1
GAIX01005024
JAA87536.1
NWSH01001222
PCG72054.1
+ More
AGBW02011133 OWR47161.1 KQ460813 KPJ12138.1 BABH01002264 BABH01002265 KQ459604 KPI92469.1 GEZM01092737 JAV56476.1 GDQN01000671 JAT90383.1 KQ971338 KYB28019.1 KYB28018.1 GALX01004430 JAB64036.1 NEVH01027072 PNF13873.1 GANO01003235 JAB56636.1 CVRI01000052 CRK99736.1 UFQT01000375 SSX23695.1 CH477432 EAT41038.1 JXUM01089177 KQ563750 KXJ73388.1 GAPW01001689 JAC11909.1 APCN01000612 AAAB01008859 EAA07843.4 AXCN02000194 KB631647 ERL84996.1 GDAI01002352 JAI15251.1 AXCM01000884 APGK01052144 APGK01052145 APGK01052146 APGK01052147 APGK01052148 KB741210 ENN72834.1 GGFM01002151 MBW22902.1 GECU01031015 JAS76691.1 ATLV01015598 ATLV01015599 ATLV01015600 KE525023 KFB40513.1 GGFM01002783 MBW23534.1 GGFJ01006512 MBW55653.1 GFTR01005445 JAW10981.1 GECZ01023948 JAS45821.1 ADMH02002131 ETN58498.1 GGFJ01005477 MBW54618.1 GGFJ01005468 MBW54609.1 KK852446 KDR23747.1 GDKW01002330 JAI54265.1 GBBI01002633 JAC16079.1 GGFK01005901 MBW39222.1 GEDC01009360 JAS27938.1 GALA01001630 JAA93222.1 GFDL01002796 JAV32249.1 GBXI01009343 GBXI01004216 JAD04949.1 JAD10076.1 GAKP01019650 GAKP01019649 JAC39303.1 GDHF01023275 JAI29039.1 GAMC01009930 JAB96625.1 GDHC01016027 JAQ02602.1 GBHO01035136 JAG08468.1 GBRD01010696 JAG55128.1 CP012528 ALC48875.1 ADTU01015225 ADTU01015226 KK107314 QOIP01000012 EZA52843.1 RLU16274.1 ACPB03015643 DS235286 EEB14434.1 KZ288282 PBC29527.1 JR040746 AEY59273.1 JRES01000981 KNC26526.1 CCAG010011817 JXJN01008939 KA648141 AFP62770.1 CH933810 EDW06664.1 CH916371 EDV91809.1 CH940653 EDW62642.2 LBMM01000179 KMR04592.1 KQ976574 KYM80219.1 KQ435727 KOX77862.1 GL440430 EFN65863.1 AE014298 AGB95019.1 NNAY01003442 OXU19409.1 GGMR01000546 MBY13165.1 GBYB01009565 JAG79332.1 CH379064 EAL31591.3 GFDF01003490 JAV10594.1 AJWK01008213 CM000162 EDX01420.1 KQ981625 KYN39135.1 CH964239 EDW82319.1 CH954183 EDV45566.1
AGBW02011133 OWR47161.1 KQ460813 KPJ12138.1 BABH01002264 BABH01002265 KQ459604 KPI92469.1 GEZM01092737 JAV56476.1 GDQN01000671 JAT90383.1 KQ971338 KYB28019.1 KYB28018.1 GALX01004430 JAB64036.1 NEVH01027072 PNF13873.1 GANO01003235 JAB56636.1 CVRI01000052 CRK99736.1 UFQT01000375 SSX23695.1 CH477432 EAT41038.1 JXUM01089177 KQ563750 KXJ73388.1 GAPW01001689 JAC11909.1 APCN01000612 AAAB01008859 EAA07843.4 AXCN02000194 KB631647 ERL84996.1 GDAI01002352 JAI15251.1 AXCM01000884 APGK01052144 APGK01052145 APGK01052146 APGK01052147 APGK01052148 KB741210 ENN72834.1 GGFM01002151 MBW22902.1 GECU01031015 JAS76691.1 ATLV01015598 ATLV01015599 ATLV01015600 KE525023 KFB40513.1 GGFM01002783 MBW23534.1 GGFJ01006512 MBW55653.1 GFTR01005445 JAW10981.1 GECZ01023948 JAS45821.1 ADMH02002131 ETN58498.1 GGFJ01005477 MBW54618.1 GGFJ01005468 MBW54609.1 KK852446 KDR23747.1 GDKW01002330 JAI54265.1 GBBI01002633 JAC16079.1 GGFK01005901 MBW39222.1 GEDC01009360 JAS27938.1 GALA01001630 JAA93222.1 GFDL01002796 JAV32249.1 GBXI01009343 GBXI01004216 JAD04949.1 JAD10076.1 GAKP01019650 GAKP01019649 JAC39303.1 GDHF01023275 JAI29039.1 GAMC01009930 JAB96625.1 GDHC01016027 JAQ02602.1 GBHO01035136 JAG08468.1 GBRD01010696 JAG55128.1 CP012528 ALC48875.1 ADTU01015225 ADTU01015226 KK107314 QOIP01000012 EZA52843.1 RLU16274.1 ACPB03015643 DS235286 EEB14434.1 KZ288282 PBC29527.1 JR040746 AEY59273.1 JRES01000981 KNC26526.1 CCAG010011817 JXJN01008939 KA648141 AFP62770.1 CH933810 EDW06664.1 CH916371 EDV91809.1 CH940653 EDW62642.2 LBMM01000179 KMR04592.1 KQ976574 KYM80219.1 KQ435727 KOX77862.1 GL440430 EFN65863.1 AE014298 AGB95019.1 NNAY01003442 OXU19409.1 GGMR01000546 MBY13165.1 GBYB01009565 JAG79332.1 CH379064 EAL31591.3 GFDF01003490 JAV10594.1 AJWK01008213 CM000162 EDX01420.1 KQ981625 KYN39135.1 CH964239 EDW82319.1 CH954183 EDV45566.1
Proteomes
UP000283053
UP000218220
UP000007151
UP000053240
UP000005204
UP000053268
+ More
UP000007266 UP000235965 UP000183832 UP000008820 UP000069940 UP000249989 UP000075901 UP000075882 UP000075903 UP000075840 UP000075902 UP000007062 UP000076407 UP000075885 UP000075900 UP000075920 UP000075881 UP000075886 UP000075884 UP000030742 UP000075883 UP000019118 UP000076408 UP000075880 UP000030765 UP000000673 UP000027135 UP000092553 UP000005205 UP000069272 UP000053097 UP000279307 UP000015103 UP000009046 UP000242457 UP000005203 UP000037069 UP000092444 UP000092445 UP000078200 UP000092460 UP000095300 UP000095301 UP000009192 UP000001070 UP000008792 UP000036403 UP000078540 UP000053105 UP000000311 UP000000803 UP000002358 UP000215335 UP000091820 UP000001819 UP000092461 UP000002282 UP000078541 UP000007798 UP000008711
UP000007266 UP000235965 UP000183832 UP000008820 UP000069940 UP000249989 UP000075901 UP000075882 UP000075903 UP000075840 UP000075902 UP000007062 UP000076407 UP000075885 UP000075900 UP000075920 UP000075881 UP000075886 UP000075884 UP000030742 UP000075883 UP000019118 UP000076408 UP000075880 UP000030765 UP000000673 UP000027135 UP000092553 UP000005205 UP000069272 UP000053097 UP000279307 UP000015103 UP000009046 UP000242457 UP000005203 UP000037069 UP000092444 UP000092445 UP000078200 UP000092460 UP000095300 UP000095301 UP000009192 UP000001070 UP000008792 UP000036403 UP000078540 UP000053105 UP000000311 UP000000803 UP000002358 UP000215335 UP000091820 UP000001819 UP000092461 UP000002282 UP000078541 UP000007798 UP000008711
PRIDE
Interpro
ProteinModelPortal
A0A3S2TP34
S4P6I4
A0A2A4JKD1
A0A212F0D3
A0A0N1IP27
H9JG94
+ More
A0A194PIB7 A0A1Y1KAH8 A0A1E1WTM4 A0A139WJ97 A0A139WJ27 V5I8M4 A0A2J7PC04 U5ESI1 A0A1J1IHQ8 A0A336M0C6 Q172Q9 A0A182GUG2 A0A023ETZ5 A0A182SL00 A0A182LFT5 A0A182VC56 A0A182HU79 A0A182TXW6 Q7QD97 A0A182WRJ7 A0A182P5T5 A0A182RNR3 A0A182W064 A0A182K945 A0A182QRB0 A0A182N640 U4U321 A0A0K8TM88 A0A182LZF5 N6SYH4 A0A182Y4A7 A0A2M3Z305 A0A1B6HPW0 A0A182IYB7 A0A084VRB9 A0A2M3Z4U8 A0A2M4BRP9 A0A224XSN3 A0A1B6F6T4 W5J514 A0A2M4BPA6 A0A2M4BNE6 A0A067RIX0 A0A0P4VHV2 A0A023F4Z1 A0A2M4AES3 A0A1B6DQJ0 T1E205 A0A1Q3FXE4 A0A0A1XGK8 A0A034VCN7 A0A0K8UQR5 W8B648 A0A146L607 A0A0A9WPQ4 A0A0K8SPF7 A0A0M4F8V1 A0A158NH83 A0A182FRE8 A0A026WA38 T1HXL5 E0VM28 A0A2A3ECR3 V9IDS1 A0A088AHM0 A0A0L0C2P0 A0A1B0FCH7 A0A1A9Z6D1 A0A1A9USZ9 A0A1B0B611 A0A1I8PX97 T1PH91 A0A1I8MLI2 B4L1E3 B4JM48 B4M775 A0A0J7L9M2 A0A195B7P7 A0A0N0BIL8 E2AL25 M9PGS9 K7IXY2 A0A232EM24 A0A2S2N7N8 A0A1A9X5M8 A0A0C9RMJ6 Q29I15 A0A1L8DVX9 A0A1B0CDT6 B4Q1B3 A0A195FEM8 B4NBW0 B3P8W1
A0A194PIB7 A0A1Y1KAH8 A0A1E1WTM4 A0A139WJ97 A0A139WJ27 V5I8M4 A0A2J7PC04 U5ESI1 A0A1J1IHQ8 A0A336M0C6 Q172Q9 A0A182GUG2 A0A023ETZ5 A0A182SL00 A0A182LFT5 A0A182VC56 A0A182HU79 A0A182TXW6 Q7QD97 A0A182WRJ7 A0A182P5T5 A0A182RNR3 A0A182W064 A0A182K945 A0A182QRB0 A0A182N640 U4U321 A0A0K8TM88 A0A182LZF5 N6SYH4 A0A182Y4A7 A0A2M3Z305 A0A1B6HPW0 A0A182IYB7 A0A084VRB9 A0A2M3Z4U8 A0A2M4BRP9 A0A224XSN3 A0A1B6F6T4 W5J514 A0A2M4BPA6 A0A2M4BNE6 A0A067RIX0 A0A0P4VHV2 A0A023F4Z1 A0A2M4AES3 A0A1B6DQJ0 T1E205 A0A1Q3FXE4 A0A0A1XGK8 A0A034VCN7 A0A0K8UQR5 W8B648 A0A146L607 A0A0A9WPQ4 A0A0K8SPF7 A0A0M4F8V1 A0A158NH83 A0A182FRE8 A0A026WA38 T1HXL5 E0VM28 A0A2A3ECR3 V9IDS1 A0A088AHM0 A0A0L0C2P0 A0A1B0FCH7 A0A1A9Z6D1 A0A1A9USZ9 A0A1B0B611 A0A1I8PX97 T1PH91 A0A1I8MLI2 B4L1E3 B4JM48 B4M775 A0A0J7L9M2 A0A195B7P7 A0A0N0BIL8 E2AL25 M9PGS9 K7IXY2 A0A232EM24 A0A2S2N7N8 A0A1A9X5M8 A0A0C9RMJ6 Q29I15 A0A1L8DVX9 A0A1B0CDT6 B4Q1B3 A0A195FEM8 B4NBW0 B3P8W1
Ontologies
PANTHER
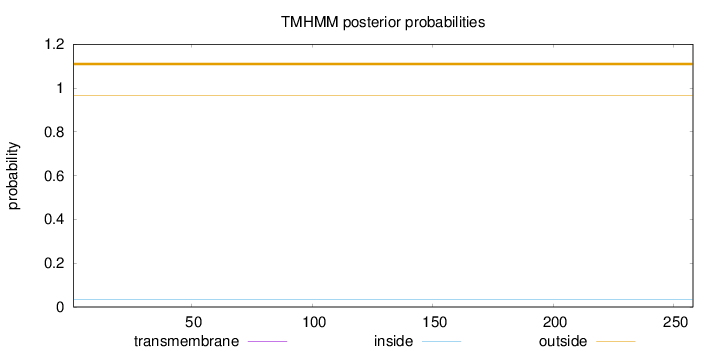
Topology
Length:
258
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00113
Exp number, first 60 AAs:
0.00102
Total prob of N-in:
0.03384
outside
1 - 258
Population Genetic Test Statistics
Pi
248.143741
Theta
172.613407
Tajima's D
1.486151
CLR
0.214939
CSRT
0.782860856957152
Interpretation
Uncertain
