Gene
KWMTBOMO11038 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008538
Annotation
arylalkylamine_N-acetyltransferase_[Bombyx_mori]
Full name
Dopamine N-acetyltransferase
Alternative Name
Arylalkylamine N-acetyltransferase
Location in the cell
Cytoplasmic Reliability : 2.091
Sequence
CDS
ATGCTCGGAGACGAACTGCCGGCGCAGCTTTCGAGCTTGGAGATCCGGCCTCGCAAAATGTCCGTTCCAGCTTACACCATACAGAGGCTCACGTACAACGATCGGGACCTAGTACTGAAATTCCTCAGAAGATTCTTCTTTCGCGATGAGCCAATGAATCTCGCCGTCAATCTGCTTGAAACACCAGAGTCAAGATGCACTGAGCTCGATGACTATGCGGCAGCTACCTTGAGCGATGGAGTTTCAGTCGCCGCTGTTGATGAGAACGGCGACTACGTAGGAGTCATCATTAACGGGATTGTGAGGAGAGAGGAAGTAGACTACACGGACAAGTCTGAAGACTGCCCCAATCCAAAATTCCGACGGATCCTGAAAGTACTGGGTCACTTGGACCGCGAAGCTCGCATCTGGGACAAACTGCCGGAGACTTGTGACTCTGTTCTCGAGATCAGGATAGCGTCTACACACTCGTCTTGGCGCGGCAGAGGACTGATGAGGGTGCTGTGTGAGGAAGCGGAACGTCTGGCCAAAGCCATGGGAGCCGGAGCTCTTAGAATGGATACAACGTCAGCTTTCTCTGCGGCCGCTGCGGAGAGGCTCAATTACAAGATGGCGTTTGGGGTTCGGTATGCTGATTTGCCGTACGCACCTCAACCTGAAGCACCACACCTTGAAGCCAGGGTCTACATTAAGGAGCTGTAA
Protein
MLGDELPAQLSSLEIRPRKMSVPAYTIQRLTYNDRDLVLKFLRRFFFRDEPMNLAVNLLETPESRCTELDDYAAATLSDGVSVAAVDENGDYVGVIINGIVRREEVDYTDKSEDCPNPKFRRILKVLGHLDREARIWDKLPETCDSVLEIRIASTHSSWRGRGLMRVLCEEAERLAKAMGAGALRMDTTSAFSAAAAERLNYKMAFGVRYADLPYAPQPEAPHLEARVYIKEL
Summary
Description
Catalyzes N-acetylation of tryptamine, tyramine, dopamine, serotonin and octopamine. Is not essential for sclerotization.
Catalytic Activity
2-arylethylamine + acetyl-CoA = CoA + H(+) + N-acetyl-2-arylethylamine
Biophysicochemical Properties
0.19 mM for tryptamine (in transfected COS-7 cell extracts)
0.89 mM for tryptamine (using purified enzyme from flies after hydroxylapatite chromatography)
1.15 mM for dopamine (in transfected COS-7 cell extracts)
0.97 mM for dopamine (using purified enzyme from flies after hydroxylapatite chromatography)
1.62 mM for serotonin (in transfected COS-7 cell extracts)
0.91 mM for serotonin (using purified enzyme from flies after hydroxylapatite chromatography)
0.89 mM for tryptamine (using purified enzyme from flies after hydroxylapatite chromatography)
1.15 mM for dopamine (in transfected COS-7 cell extracts)
0.97 mM for dopamine (using purified enzyme from flies after hydroxylapatite chromatography)
1.62 mM for serotonin (in transfected COS-7 cell extracts)
0.91 mM for serotonin (using purified enzyme from flies after hydroxylapatite chromatography)
Keywords
3D-structure
Acyltransferase
Alternative splicing
Complete proteome
Direct protein sequencing
Melatonin biosynthesis
Reference proteome
Transferase
Feature
chain Dopamine N-acetyltransferase
splice variant In isoform A.
splice variant In isoform A.
Uniprot
A0EM56
H9JG91
A2SZ52
D5LPS6
A0A194PIC1
A0A0N1PG91
+ More
A0A0G3VGS2 A0A1E1VZG3 A0A2A4JDQ1 A0A3S2M5D3 A0A2H1VQT5 A0A2H1VQM5 A0A1B6HCH3 A0A1B6G7D1 A0A1B6FNE4 A0A2J7PVW3 A0A194PHE6 A0A0N1I6F1 A0A1Y1K969 A0A1B6EAE7 A0A067RIR6 Q76EI8 A0A0F6PL61 A0A067RHB4 H9JG92 R4UKH6 A0A146LGW6 A0A0A9ZC44 A0A146KRC8 A0A0V0G2Q8 A0A195DYD3 A0A224XX80 A0A0P4VME3 R4FQP0 A0A195B6S7 T1IG91 A0A195FFY7 A0A151XCX3 A0A158NGU6 F4WZQ5 A0A146B0I0 U4U4U9 A0A1Y1K9F0 A0A1Y1K8M0 E0VM27 A0A026W9X1 A0A2R7WYS7 A0A1Y1L7Z9 E2AL26 A0A1Y1L398 A0A1Y1L2X8 V9IEZ0 E2BVK3 A0A0J7L9R0 A0A2A3EEC9 V9IDN1 D2A0F0 A0A0L7RID8 A0A1J0AHX8 A0A139WJ51 A0A0Q9W5M3 A0A088AHR1 B4LJV2 A0A0Q9X7Z8 A0A0Q9X6F7 A0A1B0A475 T1PBJ7 B4NMS2 B4KNV5 A0A0M3QUJ6 D3TLI5 A0A023F7Y2 A0A1I8NFY9 A0A1W4UM97 A0A1B0FG71 A0A2S2QUJ8 A0A1W4V1A5 A0A3B0JNQ8 Q94521-2 B3MDN7 A0A3B0JVX0 Q94521 R4V140 A0A154NZL6 R4V906 C4WSQ4 A0A1B0AWQ3 A0A0Q5VPC8
A0A0G3VGS2 A0A1E1VZG3 A0A2A4JDQ1 A0A3S2M5D3 A0A2H1VQT5 A0A2H1VQM5 A0A1B6HCH3 A0A1B6G7D1 A0A1B6FNE4 A0A2J7PVW3 A0A194PHE6 A0A0N1I6F1 A0A1Y1K969 A0A1B6EAE7 A0A067RIR6 Q76EI8 A0A0F6PL61 A0A067RHB4 H9JG92 R4UKH6 A0A146LGW6 A0A0A9ZC44 A0A146KRC8 A0A0V0G2Q8 A0A195DYD3 A0A224XX80 A0A0P4VME3 R4FQP0 A0A195B6S7 T1IG91 A0A195FFY7 A0A151XCX3 A0A158NGU6 F4WZQ5 A0A146B0I0 U4U4U9 A0A1Y1K9F0 A0A1Y1K8M0 E0VM27 A0A026W9X1 A0A2R7WYS7 A0A1Y1L7Z9 E2AL26 A0A1Y1L398 A0A1Y1L2X8 V9IEZ0 E2BVK3 A0A0J7L9R0 A0A2A3EEC9 V9IDN1 D2A0F0 A0A0L7RID8 A0A1J0AHX8 A0A139WJ51 A0A0Q9W5M3 A0A088AHR1 B4LJV2 A0A0Q9X7Z8 A0A0Q9X6F7 A0A1B0A475 T1PBJ7 B4NMS2 B4KNV5 A0A0M3QUJ6 D3TLI5 A0A023F7Y2 A0A1I8NFY9 A0A1W4UM97 A0A1B0FG71 A0A2S2QUJ8 A0A1W4V1A5 A0A3B0JNQ8 Q94521-2 B3MDN7 A0A3B0JVX0 Q94521 R4V140 A0A154NZL6 R4V906 C4WSQ4 A0A1B0AWQ3 A0A0Q5VPC8
EC Number
2.3.1.87
Pubmed
EMBL
DQ256382
ABB70231.1
BABH01002257
BABH01002258
DQ372910
ABD17803.1
+ More
GU953216 ADF43200.1 KQ459604 KPI92474.1 KQ460813 KPJ12134.1 KP657626 AKL78851.1 GDQN01010919 JAT80135.1 NWSH01001851 PCG69916.1 RSAL01000035 RVE51340.1 ODYU01003860 SOQ43146.1 SOQ43145.1 GECU01035326 GECU01017712 JAS72380.1 JAS89994.1 GECZ01011536 JAS58233.1 GECZ01018069 JAS51700.1 NEVH01020940 PNF20477.1 KPI92473.1 KPJ12135.1 GEZM01094047 JAV55976.1 GEDC01028707 GEDC01002464 JAS08591.1 JAS34834.1 KK852446 KDR23746.1 AB106562 BAC87874.1 KJ865242 AJS19086.1 KK852473 KDR23147.1 BABH01002260 KC740795 AGM32619.1 GDHC01012427 GDHC01005399 JAQ06202.1 JAQ13230.1 GBHO01004224 JAG39380.1 GDHC01020837 JAP97791.1 GECL01003704 JAP02420.1 KQ980107 KYN17712.1 GFTR01003767 JAW12659.1 GDKW01003121 JAI53474.1 GAHY01000294 JAA77216.1 KQ976574 KYM80218.1 ACPB03006810 KQ981625 KYN39137.1 KQ982294 KYQ58226.1 ADTU01015227 ADTU01015228 GL888480 EGI60330.1 KX023898 AMW91815.1 KB631647 ERL84995.1 GEZM01093935 JAV56066.1 GEZM01093936 JAV56065.1 DS235286 EEB14433.1 KK107314 QOIP01000012 EZA52845.1 RLU16273.1 KK856027 PTY24677.1 GEZM01066220 GEZM01066217 JAV68055.1 GL440430 EFN65864.1 GEZM01066218 JAV68054.1 GEZM01066219 JAV68053.1 JR040667 AEY59232.1 GL450875 EFN80273.1 LBMM01000179 KMR04593.1 KZ288282 PBC29526.1 JR040668 AEY59233.1 KQ971338 EFA02503.2 KQ414584 KOC70563.1 KX812753 APB53910.1 KYB28020.1 CH940648 KRF80098.1 EDW61606.1 CH933808 KRG04412.1 CH964282 KRG00263.1 KA646044 KA647918 AFP60673.1 EDW85661.1 EDW09000.2 CP012524 ALC40789.1 EZ422287 ADD18563.1 GBBI01001364 JAC17348.1 CCAG010011116 GGMS01012191 MBY81394.1 OUUW01000001 SPP75219.1 Y07964 AE013599 AY118780 AAF47172.1 AAM50640.1 CAA69262.1 CH902619 EDV37500.1 SPP75218.1 KC626025 KC626027 KC626028 AGM38188.1 AGM38190.1 AGM38191.1 KQ434786 KZC05043.1 KC626026 AGM38189.1 ABLF02035746 AK340292 BAH70924.1 JXJN01004888 CH954179 KQS63110.1
GU953216 ADF43200.1 KQ459604 KPI92474.1 KQ460813 KPJ12134.1 KP657626 AKL78851.1 GDQN01010919 JAT80135.1 NWSH01001851 PCG69916.1 RSAL01000035 RVE51340.1 ODYU01003860 SOQ43146.1 SOQ43145.1 GECU01035326 GECU01017712 JAS72380.1 JAS89994.1 GECZ01011536 JAS58233.1 GECZ01018069 JAS51700.1 NEVH01020940 PNF20477.1 KPI92473.1 KPJ12135.1 GEZM01094047 JAV55976.1 GEDC01028707 GEDC01002464 JAS08591.1 JAS34834.1 KK852446 KDR23746.1 AB106562 BAC87874.1 KJ865242 AJS19086.1 KK852473 KDR23147.1 BABH01002260 KC740795 AGM32619.1 GDHC01012427 GDHC01005399 JAQ06202.1 JAQ13230.1 GBHO01004224 JAG39380.1 GDHC01020837 JAP97791.1 GECL01003704 JAP02420.1 KQ980107 KYN17712.1 GFTR01003767 JAW12659.1 GDKW01003121 JAI53474.1 GAHY01000294 JAA77216.1 KQ976574 KYM80218.1 ACPB03006810 KQ981625 KYN39137.1 KQ982294 KYQ58226.1 ADTU01015227 ADTU01015228 GL888480 EGI60330.1 KX023898 AMW91815.1 KB631647 ERL84995.1 GEZM01093935 JAV56066.1 GEZM01093936 JAV56065.1 DS235286 EEB14433.1 KK107314 QOIP01000012 EZA52845.1 RLU16273.1 KK856027 PTY24677.1 GEZM01066220 GEZM01066217 JAV68055.1 GL440430 EFN65864.1 GEZM01066218 JAV68054.1 GEZM01066219 JAV68053.1 JR040667 AEY59232.1 GL450875 EFN80273.1 LBMM01000179 KMR04593.1 KZ288282 PBC29526.1 JR040668 AEY59233.1 KQ971338 EFA02503.2 KQ414584 KOC70563.1 KX812753 APB53910.1 KYB28020.1 CH940648 KRF80098.1 EDW61606.1 CH933808 KRG04412.1 CH964282 KRG00263.1 KA646044 KA647918 AFP60673.1 EDW85661.1 EDW09000.2 CP012524 ALC40789.1 EZ422287 ADD18563.1 GBBI01001364 JAC17348.1 CCAG010011116 GGMS01012191 MBY81394.1 OUUW01000001 SPP75219.1 Y07964 AE013599 AY118780 AAF47172.1 AAM50640.1 CAA69262.1 CH902619 EDV37500.1 SPP75218.1 KC626025 KC626027 KC626028 AGM38188.1 AGM38190.1 AGM38191.1 KQ434786 KZC05043.1 KC626026 AGM38189.1 ABLF02035746 AK340292 BAH70924.1 JXJN01004888 CH954179 KQS63110.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000283053
UP000235965
+ More
UP000027135 UP000078492 UP000078540 UP000015103 UP000078541 UP000075809 UP000005205 UP000007755 UP000030742 UP000009046 UP000053097 UP000279307 UP000000311 UP000008237 UP000036403 UP000242457 UP000007266 UP000053825 UP000008792 UP000005203 UP000009192 UP000007798 UP000092445 UP000092553 UP000095301 UP000192221 UP000092444 UP000268350 UP000000803 UP000007801 UP000076502 UP000007819 UP000092460 UP000008711
UP000027135 UP000078492 UP000078540 UP000015103 UP000078541 UP000075809 UP000005205 UP000007755 UP000030742 UP000009046 UP000053097 UP000279307 UP000000311 UP000008237 UP000036403 UP000242457 UP000007266 UP000053825 UP000008792 UP000005203 UP000009192 UP000007798 UP000092445 UP000092553 UP000095301 UP000192221 UP000092444 UP000268350 UP000000803 UP000007801 UP000076502 UP000007819 UP000092460 UP000008711
SUPFAM
SSF55729
SSF55729
ProteinModelPortal
A0EM56
H9JG91
A2SZ52
D5LPS6
A0A194PIC1
A0A0N1PG91
+ More
A0A0G3VGS2 A0A1E1VZG3 A0A2A4JDQ1 A0A3S2M5D3 A0A2H1VQT5 A0A2H1VQM5 A0A1B6HCH3 A0A1B6G7D1 A0A1B6FNE4 A0A2J7PVW3 A0A194PHE6 A0A0N1I6F1 A0A1Y1K969 A0A1B6EAE7 A0A067RIR6 Q76EI8 A0A0F6PL61 A0A067RHB4 H9JG92 R4UKH6 A0A146LGW6 A0A0A9ZC44 A0A146KRC8 A0A0V0G2Q8 A0A195DYD3 A0A224XX80 A0A0P4VME3 R4FQP0 A0A195B6S7 T1IG91 A0A195FFY7 A0A151XCX3 A0A158NGU6 F4WZQ5 A0A146B0I0 U4U4U9 A0A1Y1K9F0 A0A1Y1K8M0 E0VM27 A0A026W9X1 A0A2R7WYS7 A0A1Y1L7Z9 E2AL26 A0A1Y1L398 A0A1Y1L2X8 V9IEZ0 E2BVK3 A0A0J7L9R0 A0A2A3EEC9 V9IDN1 D2A0F0 A0A0L7RID8 A0A1J0AHX8 A0A139WJ51 A0A0Q9W5M3 A0A088AHR1 B4LJV2 A0A0Q9X7Z8 A0A0Q9X6F7 A0A1B0A475 T1PBJ7 B4NMS2 B4KNV5 A0A0M3QUJ6 D3TLI5 A0A023F7Y2 A0A1I8NFY9 A0A1W4UM97 A0A1B0FG71 A0A2S2QUJ8 A0A1W4V1A5 A0A3B0JNQ8 Q94521-2 B3MDN7 A0A3B0JVX0 Q94521 R4V140 A0A154NZL6 R4V906 C4WSQ4 A0A1B0AWQ3 A0A0Q5VPC8
A0A0G3VGS2 A0A1E1VZG3 A0A2A4JDQ1 A0A3S2M5D3 A0A2H1VQT5 A0A2H1VQM5 A0A1B6HCH3 A0A1B6G7D1 A0A1B6FNE4 A0A2J7PVW3 A0A194PHE6 A0A0N1I6F1 A0A1Y1K969 A0A1B6EAE7 A0A067RIR6 Q76EI8 A0A0F6PL61 A0A067RHB4 H9JG92 R4UKH6 A0A146LGW6 A0A0A9ZC44 A0A146KRC8 A0A0V0G2Q8 A0A195DYD3 A0A224XX80 A0A0P4VME3 R4FQP0 A0A195B6S7 T1IG91 A0A195FFY7 A0A151XCX3 A0A158NGU6 F4WZQ5 A0A146B0I0 U4U4U9 A0A1Y1K9F0 A0A1Y1K8M0 E0VM27 A0A026W9X1 A0A2R7WYS7 A0A1Y1L7Z9 E2AL26 A0A1Y1L398 A0A1Y1L2X8 V9IEZ0 E2BVK3 A0A0J7L9R0 A0A2A3EEC9 V9IDN1 D2A0F0 A0A0L7RID8 A0A1J0AHX8 A0A139WJ51 A0A0Q9W5M3 A0A088AHR1 B4LJV2 A0A0Q9X7Z8 A0A0Q9X6F7 A0A1B0A475 T1PBJ7 B4NMS2 B4KNV5 A0A0M3QUJ6 D3TLI5 A0A023F7Y2 A0A1I8NFY9 A0A1W4UM97 A0A1B0FG71 A0A2S2QUJ8 A0A1W4V1A5 A0A3B0JNQ8 Q94521-2 B3MDN7 A0A3B0JVX0 Q94521 R4V140 A0A154NZL6 R4V906 C4WSQ4 A0A1B0AWQ3 A0A0Q5VPC8
PDB
5GI9
E-value=6.30678e-21,
Score=246
Ontologies
GO
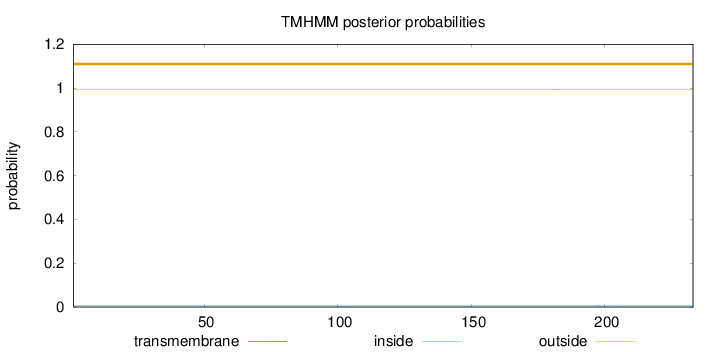
Topology
Length:
233
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00171
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00740
outside
1 - 233
Population Genetic Test Statistics
Pi
158.110198
Theta
151.420941
Tajima's D
0.503983
CLR
0.023357
CSRT
0.52122393880306
Interpretation
Possibly Positive selection
