Gene
KWMTBOMO11035
Pre Gene Modal
BGIBMGA002329
Annotation
PREDICTED:_uncharacterized_protein_LOC105254802?_partial_[Camponotus_floridanus]
Full name
ATP-dependent DNA helicase
Location in the cell
PlasmaMembrane Reliability : 2.131
Sequence
CDS
ATGCATCGTGTAAACGAGACACATCGTTTGTACGATGCGTTACAATATCCAATCATTTATTGGCAAGGACAAGACGGATACGACATCACGTTGAAGATGGTCGATCCAATTACAGGCGTATCAACGAATAAAAATCTAAGCGCAATGAATTACTATGCGTATCGTATGATGATTCGTACACATGAGGAGAATGTCATTCTGAAGTGCCGTCGGCTATTCCAGCAATTCGCTGTCGACATGTATGTCAAAGTCGAGACCGAACGTTTAGCGTTCATCCGATTCAATCAGGCAAAGCTACGATCTGAGGACTATATACACTTGCGTGATGCTATTCATTCAGATGGTGATGTTCAAAATATTGGACGTCTGACGATTCTCCCATCATCTTATATCGGAAGCCCACGCCACATGCACGAATACGCTCAAGACGCTATGACGTACGTGCGAAATTATGGAACTCCGGATTTATTTATTACGTTCACATGCAATCCGAAGTGGACGGAAATTGAACTTAAAGTTCAAAATCAAGAGATTGAGATCGGAAATGAATTCATTGTACCATATTGCCCGCTGCTATCAAGAATTTTCGAAACACATGCAAACGTTGAGAGTTGTCATTCGGCCAAATCAATCAAATATTTGTGCAAGTACGTCACAAAAGGCAGCGACATGGCTGTGTTTGGTATTGCGTCGGAAAATGCCAATGATGAAATCAGTAACTTCCAAATGGGCAGATACGTTAGTACTAATGAAGCACTGTGGCGATTATTGTCATTTCAAATTCATGAAAGATATCCCACAGTTGTACATTTAGCAGTGCATTTGGAAAATGGCCAAAGAGTTTACTTCACTGAGGCTAATGCGGCACAACGAGCTGAAAGACCACCATCGACAACATTGACTAGCTTCTTTGCAATGTGTGAAGCAGATCCATTCGCAGCGACGCTGATGTACGTTGAAATGCCCAAGTATTACACTTGGAATCAATCAACAAAGAAATTCCAACGTCGCAAACAAGGAACCCCAGTTCCAGATTGGCCACAGGTGTTTTCCACTGATGCACTAGGTCGCATGTATACTGTTCATCCTAGAAATGACGAATGTTTTTATTTGCGACTGCTGCTGGTAAATGTACGTGGACCAAAATCATTTGCGCATTTGAAAACTGTGAATGGCCACCAATGCCAAACATATCGAGAAGCATGTCAACTATTGGGTTTGCTGGAGAACGATTCTCATTGGGATTTAACACTTGCGGATTCAGTTGTTTCATCAAATGCGTACCAAATACGAACGCTGTTCGCAATTATCATCACCACATGTTTTCCTTCACAACCAATTCAGTTATGGAACAAATTCAAAGACGACATATGTGAAGATATCTTGCATCGCTTGCGCATTCAAACGAATAATCCTGACATCCAAATAACCGATGAAATCTACAATGAAGGATTGATTCTGATTGAGGATCAATGCTTGACTATTGCAAACAAGTTACTGATTGAAGTAGGAATGATTGCGCCAAATCGATCGATGCACGATGCATTCAACCAAGAATTAAATCGAGAGCTGCAATACAATGTTGATACATTGCAGGAATTCGTGCGAAATAATGTGCCGTTACTGAATGAACAGCAAAAACAAGTATACGAAACATTAATGCAAGCGGTGGACAATAATACTGGTGGTCTATTCTTCCTGGACGCACCTGGAGGAACAGGGAAAACATTTGTCATTTCATTGATTTTGGCCACTATTCGATCAAGATGTGACATAGCTTTGGCGTTAGCATCATCTGGAATTGCGGCGACTCTTCTAGACGGCGGTCGTACTGCACATTCTGCGCTTAAGTTGCCACTCAATTTAAACACAATTGATACTCCAACATGCAATATTTCCCGATCCAGTGCAATGGGAAAATTGTTGATGCAATGCAAGCTCATTGTTTGGGATGAGTGCACAATGGCACATAAGAAATCACTTGAAGCACTTAACTTCACACTGAAGGATCTTCGGCGAAATAACAACATCTTTGGTGGCTTGATGATATTGTTGGCAGGCGATTTCAGGCAGACGTTGCCAGTAATTCCCCGTGGAACGCCTGCAGATGAATTGAATGCTTGCCTGAAGGCATCACCTTTATGGAATAACGTAAAAACATTATCGCTAGCCACTAATATGAGAGTTCAACTTCAAAATGATCAAAGTGCTGCACCATTTTCCAAACAATTGTTAGCTGTTGGAAATGGAAAAGTCCCAGTTGATGCGACATCTGGATTAATTACTCTTACCAACGACTTTTGCCGATTTGTAGACTCTCAATTAGCTCTTATTGAAAATGTTTTCCCAAACATTAGTGAGAATTATCAGAATTATGCTTGGTTAAGTCAACGAGCAATTCTTGCCGCAAAGAATAATGATGTACACGCACTGAATTTCACCATTCAATCAAAAATTGCTGGCGATTTGGTGACATACAAATCCGTTGATTCCATAACAAATCCCGATGATGTAGTAAATTATCCAACGGAGTTTTTGAACTCTCTGGAGTTGCCAGGATTTCCACCACATAACTTGCAACTCAAAGTTGGTACAGAGCACTTCAATGAAACGGATAATTTGCGGACACGTATAACGCTTCGATTCAGTATTTACTTTGGATTCACTTTCATTTACTTAGAGAAGTTCAACTAA
Protein
MHRVNETHRLYDALQYPIIYWQGQDGYDITLKMVDPITGVSTNKNLSAMNYYAYRMMIRTHEENVILKCRRLFQQFAVDMYVKVETERLAFIRFNQAKLRSEDYIHLRDAIHSDGDVQNIGRLTILPSSYIGSPRHMHEYAQDAMTYVRNYGTPDLFITFTCNPKWTEIELKVQNQEIEIGNEFIVPYCPLLSRIFETHANVESCHSAKSIKYLCKYVTKGSDMAVFGIASENANDEISNFQMGRYVSTNEALWRLLSFQIHERYPTVVHLAVHLENGQRVYFTEANAAQRAERPPSTTLTSFFAMCEADPFAATLMYVEMPKYYTWNQSTKKFQRRKQGTPVPDWPQVFSTDALGRMYTVHPRNDECFYLRLLLVNVRGPKSFAHLKTVNGHQCQTYREACQLLGLLENDSHWDLTLADSVVSSNAYQIRTLFAIIITTCFPSQPIQLWNKFKDDICEDILHRLRIQTNNPDIQITDEIYNEGLILIEDQCLTIANKLLIEVGMIAPNRSMHDAFNQELNRELQYNVDTLQEFVRNNVPLLNEQQKQVYETLMQAVDNNTGGLFFLDAPGGTGKTFVISLILATIRSRCDIALALASSGIAATLLDGGRTAHSALKLPLNLNTIDTPTCNISRSSAMGKLLMQCKLIVWDECTMAHKKSLEALNFTLKDLRRNNNIFGGLMILLAGDFRQTLPVIPRGTPADELNACLKASPLWNNVKTLSLATNMRVQLQNDQSAAPFSKQLLAVGNGKVPVDATSGLITLTNDFCRFVDSQLALIENVFPNISENYQNYAWLSQRAILAAKNNDVHALNFTIQSKIAGDLVTYKSVDSITNPDDVVNYPTEFLNSLELPGFPPHNLQLKVGTEHFNETDNLRTRITLRFSIYFGFTFIYLEKFN
Summary
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Cofactor
Mg(2+)
Similarity
Belongs to the helicase family.
Uniprot
A0A3S2NR36
H9JGG9
S4PSG3
F4ZH17
A0A3S2NIC2
A0A0D6L4E4
+ More
A0A016W7D9 A0A0C2CWG5 A0A0C2G6A8 A0A0C2G6C2 A0A267GN76 A0A267F4D8 A0A267GEW9 A0A267GWL2 A0A267F5E2 A0A368GPV9 A0A0C2DC56 X1WPL7 A0A368FRT6 X1WX02 A0A267F2R0 A0A1S3DI71 J9LSB3 X1X109 A0A2C9JJ58 J9LB24 A0A267DHT9 A0A267EIS0 A0A267F6F6 A0A1U7YIN1 A0A267GBS1 A0A015KY09 A0A267GS15 A0A267DCD1 A0A267GG99 A0A267ETE1 A0A267ELF5 A0A267DIK3 A0A2H5R2Y1 A0A267ESS5 A0A2Z6RD64 A0A1I8JBS8 A0A267H554 A0A267FG92 A0A2H5U846 A0A2H5SHM7 A0A3P6E0M8 A0A1S4A6U6 R7QQQ7 A0A0C2WEL5 A0A2G5F930 K3YD59
A0A016W7D9 A0A0C2CWG5 A0A0C2G6A8 A0A0C2G6C2 A0A267GN76 A0A267F4D8 A0A267GEW9 A0A267GWL2 A0A267F5E2 A0A368GPV9 A0A0C2DC56 X1WPL7 A0A368FRT6 X1WX02 A0A267F2R0 A0A1S3DI71 J9LSB3 X1X109 A0A2C9JJ58 J9LB24 A0A267DHT9 A0A267EIS0 A0A267F6F6 A0A1U7YIN1 A0A267GBS1 A0A015KY09 A0A267GS15 A0A267DCD1 A0A267GG99 A0A267ETE1 A0A267ELF5 A0A267DIK3 A0A2H5R2Y1 A0A267ESS5 A0A2Z6RD64 A0A1I8JBS8 A0A267H554 A0A267FG92 A0A2H5U846 A0A2H5SHM7 A0A3P6E0M8 A0A1S4A6U6 R7QQQ7 A0A0C2WEL5 A0A2G5F930 K3YD59
EC Number
3.6.4.12
Pubmed
EMBL
RSAL01000338
RVE42429.1
BABH01039432
GAIX01013838
JAA78722.1
HQ009558
+ More
AEE09607.1 RSAL01001639 RVE40746.1 KE126587 EPB66060.1 JARK01000714 EYC35207.1 KN739487 KIH54222.1 KN739098 KIH54434.1 KN743176 KIH52626.1 NIVC01000229 PAA87485.1 NIVC01001445 PAA67902.1 NIVC01000366 PAA84583.1 NIVC01000111 PAA90396.1 NIVC01001359 PAA68995.1 JOJR01000111 RCN45090.1 KN726708 KIH67418.1 ABLF02016932 JOJR01000723 RCN34924.1 ABLF02041819 NIVC01001486 PAA67379.1 ABLF02024220 ABLF02041348 ABLF02020814 ABLF02049665 ABLF02018127 NIVC01004230 PAA48122.1 NIVC01002034 PAA61410.1 NIVC01001383 PAA68602.1 NIVC01000419 PAA83453.1 JEMT01022708 EXX64911.1 NIVC01000175 PAA88803.1 NIVC01004666 PAA46875.1 NIVC01000381 PAA84309.1 NIVC01001722 PAA64736.1 NIVC01001945 PAA62370.1 NIVC01003989 PAA49045.1 BDIQ01000010 GBC12419.1 NIVC01002018 NIVC01001743 NIVC01000051 PAA61561.1 PAA64516.1 PAA92464.1 BEXD01001738 GBB95559.1 NIVC01000042 PAA92669.1 NIVC01001115 PAA72069.1 BDIQ01000188 GBC50999.1 BDIQ01000091 GBC29771.1 LR031875 VDD29751.1 HG002121 CDF40068.1 KN818726 KIL54493.1 KZ305018 PIA64450.1 AGNK02004404
AEE09607.1 RSAL01001639 RVE40746.1 KE126587 EPB66060.1 JARK01000714 EYC35207.1 KN739487 KIH54222.1 KN739098 KIH54434.1 KN743176 KIH52626.1 NIVC01000229 PAA87485.1 NIVC01001445 PAA67902.1 NIVC01000366 PAA84583.1 NIVC01000111 PAA90396.1 NIVC01001359 PAA68995.1 JOJR01000111 RCN45090.1 KN726708 KIH67418.1 ABLF02016932 JOJR01000723 RCN34924.1 ABLF02041819 NIVC01001486 PAA67379.1 ABLF02024220 ABLF02041348 ABLF02020814 ABLF02049665 ABLF02018127 NIVC01004230 PAA48122.1 NIVC01002034 PAA61410.1 NIVC01001383 PAA68602.1 NIVC01000419 PAA83453.1 JEMT01022708 EXX64911.1 NIVC01000175 PAA88803.1 NIVC01004666 PAA46875.1 NIVC01000381 PAA84309.1 NIVC01001722 PAA64736.1 NIVC01001945 PAA62370.1 NIVC01003989 PAA49045.1 BDIQ01000010 GBC12419.1 NIVC01002018 NIVC01001743 NIVC01000051 PAA61561.1 PAA64516.1 PAA92464.1 BEXD01001738 GBB95559.1 NIVC01000042 PAA92669.1 NIVC01001115 PAA72069.1 BDIQ01000188 GBC50999.1 BDIQ01000091 GBC29771.1 LR031875 VDD29751.1 HG002121 CDF40068.1 KN818726 KIL54493.1 KZ305018 PIA64450.1 AGNK02004404
Proteomes
PRIDE
Interpro
ProteinModelPortal
A0A3S2NR36
H9JGG9
S4PSG3
F4ZH17
A0A3S2NIC2
A0A0D6L4E4
+ More
A0A016W7D9 A0A0C2CWG5 A0A0C2G6A8 A0A0C2G6C2 A0A267GN76 A0A267F4D8 A0A267GEW9 A0A267GWL2 A0A267F5E2 A0A368GPV9 A0A0C2DC56 X1WPL7 A0A368FRT6 X1WX02 A0A267F2R0 A0A1S3DI71 J9LSB3 X1X109 A0A2C9JJ58 J9LB24 A0A267DHT9 A0A267EIS0 A0A267F6F6 A0A1U7YIN1 A0A267GBS1 A0A015KY09 A0A267GS15 A0A267DCD1 A0A267GG99 A0A267ETE1 A0A267ELF5 A0A267DIK3 A0A2H5R2Y1 A0A267ESS5 A0A2Z6RD64 A0A1I8JBS8 A0A267H554 A0A267FG92 A0A2H5U846 A0A2H5SHM7 A0A3P6E0M8 A0A1S4A6U6 R7QQQ7 A0A0C2WEL5 A0A2G5F930 K3YD59
A0A016W7D9 A0A0C2CWG5 A0A0C2G6A8 A0A0C2G6C2 A0A267GN76 A0A267F4D8 A0A267GEW9 A0A267GWL2 A0A267F5E2 A0A368GPV9 A0A0C2DC56 X1WPL7 A0A368FRT6 X1WX02 A0A267F2R0 A0A1S3DI71 J9LSB3 X1X109 A0A2C9JJ58 J9LB24 A0A267DHT9 A0A267EIS0 A0A267F6F6 A0A1U7YIN1 A0A267GBS1 A0A015KY09 A0A267GS15 A0A267DCD1 A0A267GG99 A0A267ETE1 A0A267ELF5 A0A267DIK3 A0A2H5R2Y1 A0A267ESS5 A0A2Z6RD64 A0A1I8JBS8 A0A267H554 A0A267FG92 A0A2H5U846 A0A2H5SHM7 A0A3P6E0M8 A0A1S4A6U6 R7QQQ7 A0A0C2WEL5 A0A2G5F930 K3YD59
PDB
5FHH
E-value=8.36803e-05,
Score=113
Ontologies
GO
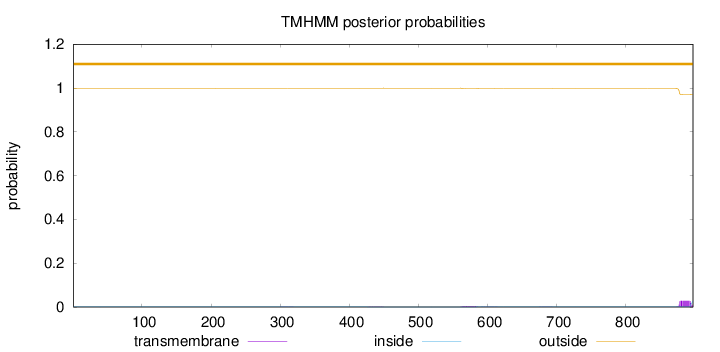
Topology
Length:
897
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.5988
Exp number, first 60 AAs:
0.00236
Total prob of N-in:
0.00091
outside
1 - 897
Population Genetic Test Statistics
Pi
17.151575
Theta
35.847552
Tajima's D
0
CLR
249.213702
CSRT
0.369281535923204
Interpretation
Uncertain
