Gene
KWMTBOMO11022
Pre Gene Modal
BGIBMGA008531
Annotation
Myb-MuvB_complex_subunit_Lin-52_[Bombyx_mori]
Full name
Protein lin-52 homolog
Location in the cell
Nuclear Reliability : 2.381
Sequence
CDS
ATGGTGAACATGGCCGCGAAACAAAATTCAATCGTTGATGACCAAGCATCATCCGATGATATTCCGTTGACATCCTTAGAGGAAAGTCTTTTCAGTTCGGAGAAGCTTGATAGAGCTTCACCAGAACTATGGCCTGAACAAATTCCGGGAGTATCGGAGTTTGCACCGATGCCAGGCACGCAGGAGCCTTCCTGGAATGAAGGTCTCACAAAACAGGACTACACTTACATGCAACAACTTGGAACACTTACAGTGAGTGGTCTCATCATGGAAGTCAAAAAACTTCATGACTTAGCTTATCAACTGGGCCTGGAAGAAGCTAAAGAGATGACCCGTGGAAAATACTTGAATATATTTGCATCAAAAAGGCAACGGTAG
Protein
MVNMAAKQNSIVDDQASSDDIPLTSLEESLFSSEKLDRASPELWPEQIPGVSEFAPMPGTQEPSWNEGLTKQDYTYMQQLGTLTVSGLIMEVKKLHDLAYQLGLEEAKEMTRGKYLNIFASKRQR
Summary
Subunit
Component of the DREAM complex.
Similarity
Belongs to the lin-52 family.
Keywords
Complete proteome
Reference proteome
Feature
chain Protein lin-52 homolog
Uniprot
Q2F5Y1
A0A212F9E8
A0A1E1VZF2
A0A0N1IH52
A0A194PID0
I4DPW4
+ More
A0A2H1WDD0 A0A1E1WLT3 E2BKR6 A0A0L7QP15 A0A067QZ79 A0A2J7RHI9 A0A0C9RDZ9 E2AD92 A0A195EU27 F4WZF9 A0A195DQB5 A0A3L8DFM4 A0A0C9RKR1 A0A151IIN7 A0A195BXX6 A0A151X246 A0A1B6CA92 A0A2A3ER71 A0A0N0BDK5 A0A088AP62 U5EWG3 A0A1A9W9N9 V5I7X8 Q17CH2 A0A1I8MHW0 A0A023ED62 A0A1I8PCK1 A0A1B6IMU9 A0A026W049 A0A1B6H2U5 A0A1Q3F8J7 A0A182GVP3 B0W8N2 A0A0A0QXK1 A0A0K8TNW4 C3YGP1 A0A1B0AYR0 A0A0L0BRL1 A0A1Y1N6A1 W4Z3C9 A0A1A9UWH0 A0A0A1WTS5 A0A1B0FEB0 A0A1B0AAX3 W8CBQ1 A0A034WRA8 A0A0K8UJ54 A0A1S3HS93 A0A1S3HS91 V4A0L4 A0A087T2J3 A0A3R7M5E4 A0A2I4BJI8 D2A063 R7TWB3 Q4RLT3 S4RF08 T1FXH7 A0A3Q2VUC5 A0A3Q4G764 A0A3P8NAH1 A0A3P9BQD1 E0V9C9 A7RGP2 A0A0P4WDB0 A0A0P4W5Y2 A0A2B4SK19 B9EPT6 A0A3Q2ZHP4 A0A3M6UK18 G3NQX8
A0A2H1WDD0 A0A1E1WLT3 E2BKR6 A0A0L7QP15 A0A067QZ79 A0A2J7RHI9 A0A0C9RDZ9 E2AD92 A0A195EU27 F4WZF9 A0A195DQB5 A0A3L8DFM4 A0A0C9RKR1 A0A151IIN7 A0A195BXX6 A0A151X246 A0A1B6CA92 A0A2A3ER71 A0A0N0BDK5 A0A088AP62 U5EWG3 A0A1A9W9N9 V5I7X8 Q17CH2 A0A1I8MHW0 A0A023ED62 A0A1I8PCK1 A0A1B6IMU9 A0A026W049 A0A1B6H2U5 A0A1Q3F8J7 A0A182GVP3 B0W8N2 A0A0A0QXK1 A0A0K8TNW4 C3YGP1 A0A1B0AYR0 A0A0L0BRL1 A0A1Y1N6A1 W4Z3C9 A0A1A9UWH0 A0A0A1WTS5 A0A1B0FEB0 A0A1B0AAX3 W8CBQ1 A0A034WRA8 A0A0K8UJ54 A0A1S3HS93 A0A1S3HS91 V4A0L4 A0A087T2J3 A0A3R7M5E4 A0A2I4BJI8 D2A063 R7TWB3 Q4RLT3 S4RF08 T1FXH7 A0A3Q2VUC5 A0A3Q4G764 A0A3P8NAH1 A0A3P9BQD1 E0V9C9 A7RGP2 A0A0P4WDB0 A0A0P4W5Y2 A0A2B4SK19 B9EPT6 A0A3Q2ZHP4 A0A3M6UK18 G3NQX8
Pubmed
EMBL
DQ311291
ABD36236.1
AGBW02009596
OWR50350.1
GDQN01010995
GDQN01003765
+ More
JAT80059.1 JAT87289.1 KQ459718 KPJ20238.1 KQ459604 KPI92484.1 AK403811 BAM19954.1 ODYU01007879 SOQ51037.1 GDQN01003061 JAT87993.1 GL448819 EFN83764.1 KQ414842 KOC60360.1 KK853083 KDR11634.1 NEVH01003741 PNF40301.1 GBYB01005196 JAG74963.1 GL438685 EFN68607.1 KQ981979 KYN31407.1 GL888479 EGI60403.1 KQ980612 KYN15105.1 QOIP01000009 RLU19091.1 GBYB01007501 JAG77268.1 KQ977474 KYN02512.1 KQ976398 KYM92796.1 KQ982585 KYQ54355.1 GEDC01026891 JAS10407.1 KZ288193 PBC34004.1 KQ435859 KOX70592.1 GANO01002962 JAB56909.1 GALX01005660 JAB62806.1 CH477308 EAT44049.1 GAPW01006230 JAC07368.1 GECU01019460 JAS88246.1 KK107519 EZA49392.1 GECZ01000788 JAS68981.1 GFDL01011188 JAV23857.1 JXUM01091495 KQ563919 KXJ73067.1 DS231859 EDS39082.1 KM267673 AIU94799.1 GDAI01001782 JAI15821.1 GG666512 EEN60551.1 JXJN01005924 JRES01001467 KNC22677.1 GEZM01011800 JAV93432.1 AAGJ04163243 GBXI01011818 JAD02474.1 CCAG010000559 GAMC01000289 JAC06267.1 GAKP01002065 JAC56887.1 GDHF01025718 JAI26596.1 KB202990 ESO86806.1 KK113093 KFM59332.1 QCYY01002031 ROT73359.1 KQ971338 EFA01741.1 AMQN01011912 KB309179 ELT95270.1 CAAE01015019 AMQM01000166 KB095811 ESO12428.1 DS234991 EEB09985.1 DS469509 EDO49448.1 GDRN01098309 JAI58966.1 GDRN01098307 GDRN01098302 JAI58967.1 LSMT01000076 PFX28832.1 BT057661 ACM09533.1 RCHS01001388 RMX53874.1
JAT80059.1 JAT87289.1 KQ459718 KPJ20238.1 KQ459604 KPI92484.1 AK403811 BAM19954.1 ODYU01007879 SOQ51037.1 GDQN01003061 JAT87993.1 GL448819 EFN83764.1 KQ414842 KOC60360.1 KK853083 KDR11634.1 NEVH01003741 PNF40301.1 GBYB01005196 JAG74963.1 GL438685 EFN68607.1 KQ981979 KYN31407.1 GL888479 EGI60403.1 KQ980612 KYN15105.1 QOIP01000009 RLU19091.1 GBYB01007501 JAG77268.1 KQ977474 KYN02512.1 KQ976398 KYM92796.1 KQ982585 KYQ54355.1 GEDC01026891 JAS10407.1 KZ288193 PBC34004.1 KQ435859 KOX70592.1 GANO01002962 JAB56909.1 GALX01005660 JAB62806.1 CH477308 EAT44049.1 GAPW01006230 JAC07368.1 GECU01019460 JAS88246.1 KK107519 EZA49392.1 GECZ01000788 JAS68981.1 GFDL01011188 JAV23857.1 JXUM01091495 KQ563919 KXJ73067.1 DS231859 EDS39082.1 KM267673 AIU94799.1 GDAI01001782 JAI15821.1 GG666512 EEN60551.1 JXJN01005924 JRES01001467 KNC22677.1 GEZM01011800 JAV93432.1 AAGJ04163243 GBXI01011818 JAD02474.1 CCAG010000559 GAMC01000289 JAC06267.1 GAKP01002065 JAC56887.1 GDHF01025718 JAI26596.1 KB202990 ESO86806.1 KK113093 KFM59332.1 QCYY01002031 ROT73359.1 KQ971338 EFA01741.1 AMQN01011912 KB309179 ELT95270.1 CAAE01015019 AMQM01000166 KB095811 ESO12428.1 DS234991 EEB09985.1 DS469509 EDO49448.1 GDRN01098309 JAI58966.1 GDRN01098307 GDRN01098302 JAI58967.1 LSMT01000076 PFX28832.1 BT057661 ACM09533.1 RCHS01001388 RMX53874.1
Proteomes
UP000007151
UP000053240
UP000053268
UP000008237
UP000053825
UP000027135
+ More
UP000235965 UP000000311 UP000078541 UP000007755 UP000078492 UP000279307 UP000078542 UP000078540 UP000075809 UP000242457 UP000053105 UP000005203 UP000091820 UP000008820 UP000095301 UP000095300 UP000053097 UP000069940 UP000249989 UP000002320 UP000001554 UP000092460 UP000037069 UP000007110 UP000078200 UP000092444 UP000092445 UP000085678 UP000030746 UP000054359 UP000283509 UP000192220 UP000007266 UP000014760 UP000007303 UP000245300 UP000015101 UP000264840 UP000261580 UP000265100 UP000265160 UP000009046 UP000001593 UP000225706 UP000264800 UP000275408 UP000007635
UP000235965 UP000000311 UP000078541 UP000007755 UP000078492 UP000279307 UP000078542 UP000078540 UP000075809 UP000242457 UP000053105 UP000005203 UP000091820 UP000008820 UP000095301 UP000095300 UP000053097 UP000069940 UP000249989 UP000002320 UP000001554 UP000092460 UP000037069 UP000007110 UP000078200 UP000092444 UP000092445 UP000085678 UP000030746 UP000054359 UP000283509 UP000192220 UP000007266 UP000014760 UP000007303 UP000245300 UP000015101 UP000264840 UP000261580 UP000265100 UP000265160 UP000009046 UP000001593 UP000225706 UP000264800 UP000275408 UP000007635
PRIDE
Pfam
PF10044 LIN52
Interpro
IPR018737
DREAM_LIN52
ProteinModelPortal
Q2F5Y1
A0A212F9E8
A0A1E1VZF2
A0A0N1IH52
A0A194PID0
I4DPW4
+ More
A0A2H1WDD0 A0A1E1WLT3 E2BKR6 A0A0L7QP15 A0A067QZ79 A0A2J7RHI9 A0A0C9RDZ9 E2AD92 A0A195EU27 F4WZF9 A0A195DQB5 A0A3L8DFM4 A0A0C9RKR1 A0A151IIN7 A0A195BXX6 A0A151X246 A0A1B6CA92 A0A2A3ER71 A0A0N0BDK5 A0A088AP62 U5EWG3 A0A1A9W9N9 V5I7X8 Q17CH2 A0A1I8MHW0 A0A023ED62 A0A1I8PCK1 A0A1B6IMU9 A0A026W049 A0A1B6H2U5 A0A1Q3F8J7 A0A182GVP3 B0W8N2 A0A0A0QXK1 A0A0K8TNW4 C3YGP1 A0A1B0AYR0 A0A0L0BRL1 A0A1Y1N6A1 W4Z3C9 A0A1A9UWH0 A0A0A1WTS5 A0A1B0FEB0 A0A1B0AAX3 W8CBQ1 A0A034WRA8 A0A0K8UJ54 A0A1S3HS93 A0A1S3HS91 V4A0L4 A0A087T2J3 A0A3R7M5E4 A0A2I4BJI8 D2A063 R7TWB3 Q4RLT3 S4RF08 T1FXH7 A0A3Q2VUC5 A0A3Q4G764 A0A3P8NAH1 A0A3P9BQD1 E0V9C9 A7RGP2 A0A0P4WDB0 A0A0P4W5Y2 A0A2B4SK19 B9EPT6 A0A3Q2ZHP4 A0A3M6UK18 G3NQX8
A0A2H1WDD0 A0A1E1WLT3 E2BKR6 A0A0L7QP15 A0A067QZ79 A0A2J7RHI9 A0A0C9RDZ9 E2AD92 A0A195EU27 F4WZF9 A0A195DQB5 A0A3L8DFM4 A0A0C9RKR1 A0A151IIN7 A0A195BXX6 A0A151X246 A0A1B6CA92 A0A2A3ER71 A0A0N0BDK5 A0A088AP62 U5EWG3 A0A1A9W9N9 V5I7X8 Q17CH2 A0A1I8MHW0 A0A023ED62 A0A1I8PCK1 A0A1B6IMU9 A0A026W049 A0A1B6H2U5 A0A1Q3F8J7 A0A182GVP3 B0W8N2 A0A0A0QXK1 A0A0K8TNW4 C3YGP1 A0A1B0AYR0 A0A0L0BRL1 A0A1Y1N6A1 W4Z3C9 A0A1A9UWH0 A0A0A1WTS5 A0A1B0FEB0 A0A1B0AAX3 W8CBQ1 A0A034WRA8 A0A0K8UJ54 A0A1S3HS93 A0A1S3HS91 V4A0L4 A0A087T2J3 A0A3R7M5E4 A0A2I4BJI8 D2A063 R7TWB3 Q4RLT3 S4RF08 T1FXH7 A0A3Q2VUC5 A0A3Q4G764 A0A3P8NAH1 A0A3P9BQD1 E0V9C9 A7RGP2 A0A0P4WDB0 A0A0P4W5Y2 A0A2B4SK19 B9EPT6 A0A3Q2ZHP4 A0A3M6UK18 G3NQX8
PDB
6C48
E-value=1.6157e-09,
Score=143
Ontologies
GO
PANTHER
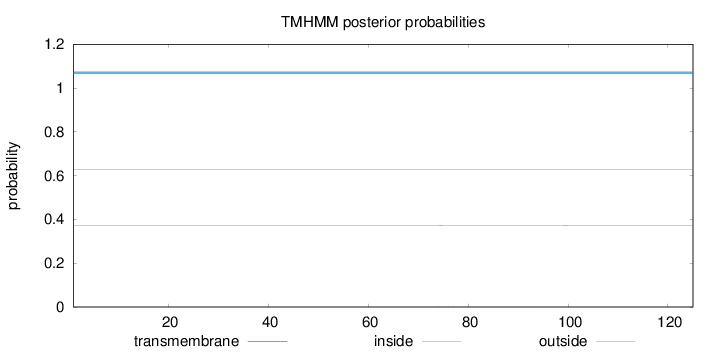
Topology
Length:
125
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01209
Exp number, first 60 AAs:
0
Total prob of N-in:
0.62787
inside
1 - 125
Population Genetic Test Statistics
Pi
169.91593
Theta
180.746235
Tajima's D
-0.133314
CLR
1.751443
CSRT
0.337283135843208
Interpretation
Uncertain
