Gene
KWMTBOMO11005
Pre Gene Modal
BGIBMGA008523
Annotation
PREDICTED:_ATP-binding_cassette_sub-family_B_member_10?_mitochondrial_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.518 Mitochondrial Reliability : 1.289
Sequence
CDS
ATGAAGTATTACACTAGCCAGATAGATGCACAAAAAAATGAAGAACCCATTGAAAATTTGAAAAATAAAACCACATTAGTCACGGGCAAGAAAATTAATGTAAAATTAAAAACTTCAGAACTAAAACGATTACTTGGACTGGCCGAACCTGAAAAATGGACATTAACTGGTGCCATTGGTTTCCTAATAGTGTCATCAAGTGTGACGATGGCCGTACCATTTTCCCTTGGCAAAGTACTTGATATAATATACAATAGTACAAGTGACTTAGCTGCTGCTAGAGAAAAGCTCGATGCATTATGTCTGATGCTCTGTGGGGTATTCTTAATAGGAGGCCTGTGTAACTTTGGAAGAGTTTACCTGATGTCCATATCTGGGCAAAGAATGACTCAGGCTCTCCGGAAACAAGTTTATGGTGCGATTTTAAAACAAGAAACGGCGTGGTTTGGAAGAACATCAACAGGGGAATTAGTTAACAGATTATCTGCAGATACACAGCTTGTGGGCCGTAATCTCAGTCAAAATGTTAGCGACGGCCTTAGGTCTTTGTTTATGGTTGGAGCTGGAACAGGAATGATGTTCTACATGTCGCCTTCATTGGCTTTGATCGGACTGTGTGTGGTACCGCCTGTTTCAATGTTGGCTGCCATATATGGACGGTTCGTCCGTGGGATCACAAGGGAGTTGCAAGATTCACTTGCGGAAACGAGCGAACTTGCTGAAGAAAAAATATCAAACATAAAGACTGTGAAAGCATTTAGCAAGGAGCCTAAGGAATGCGAATCTTATGCAAAGAGGATTGAAAATTTACTAAATTTGGCTTACAAGGAGTCATTAGCGGTCGGCAGTTTCTACGGTTTGACTGGCCTCGCGGGAAACACTATAATAATATTAGTTTTATATTACGGAGGAGGCATGGTGGCGACAGAGCAACTCACAGTCGGTAACTTGACATCGTTCCTACTGTACGCTGCGTATGTGGGGATCAGTATTGGTGGACTGAGTAGTTTCTACACGGAACTAAATAAGGGCATGGGAGCCGCGACCAGACTGTGGGAGATCATTGATAGGGAGCCCAAAATTCCTGTCACAGGAGGCTTAATACCGAAAGAGAAACCGAACGGCGAAATAATATTAGAAAAAGTACATTTCTCCTACAACGATGCGCCTCTAATACGAGGACTGGATCTTCATTTGAGACCCGGGAAGTCAGTTGCGCTTGTCGGTAGATCTGGCTGCGGGAAGAGCACCATAGCATCGCTTATATTGAGACTCTACGACCCGGATAAGGGTAGAGTTTTACTCGATGGAGTCGACATAAGGGAATTAGATCCGATGTGGTTGAGAAGTCTGATCGGCTACGTTGGTCAAGAACCGGTGTTGTTCAGTGGAACAGTCAAGGAAAATATTTTGTACGGAGCTTTAGATGAAAACGAAGAAGTACAAAATGAAAAGGACGAGCCAGCGTGGATGGTGGCAGCTCGCACTGCTCATCTGCACGAGCTAGTGACCGGCACCCGCCAAGGCTGGGACAGAGAGGTAGGCGCCGGCGGTAGTCAGCTGAGCGGGGGACAGAAACAAAGGGTGGCCATTGCACGGGCCATTGTTAAGAATCCGAAAATCTTGATCCTCGACGAAGCCACATCAGCGCTAGATACGTATTCGGAGTACTTAGTTGATAAAGCTTTAAAAGAGATCAGTAAGTCTAGAACGGTTCTGACCATAGCGCACAGGTTGAGCACGATACAGTCCGCGGACGAGGTCGCTGTGCTCGAGAATGGAGTTGTAGTTGAGACGGGACCTTACGCCGAGCTAGTAGCCAAACCGGACGGCCTCTTTAGGGAGTTGATAACGCATCAAACGTTTACCGCGAAATCGAGAGACAAAAAGGATATGCAGAAAAGCGTTTTGAGGAGGGACTTTGCTACACGTACCGCAACTGTGTCGGACAGTAAGTGCTTGAGTTCTCCGTGGGGTGAAATTCAAATTGGAAAAGAAACACTCACGGATCACGTCTTCGGTGAGGTGGAGTCATGGGCGGACAAACCATCTGTGACTTGCGGCTCATCGGGTCGTTCATACGACTACGGTATGATGCGGATGATGGTGGAACGTTGCGCCAATGCTCTCGTCGGTCATCTGAAAATGAAGCCAGGCGAGAAAATAGGCCTTATCCTGCCGAACATTCCCGAATTTGTCGTCCTCATCCATGGTGCTATGAAGGCGGGACTTGTGGTGACATTTGCGAATCCTCTGTACACTGCAGTTGAAATACAACGGCAGTTCAATGACTGTGGCGTGAAAGCAATAGCCACTATTGAGATGTTCATGCCGGTGGCCCAGGAAGTCAGCAAGTCACTGAAAGATTACAAGGGAACCATTTGGGTTGGTGGCGAAGATGATAAAGCAAAAGGTATCCACGGCCTAAGGTCATTGTTAATGGCGGATCACAAAGCGGATCTTCCCACATTGAACTGCGACGATGTGTGTTTGATACCGTACTCCAGCGGTACGACTGGAATGCCCAAAGGCGTCATGCTCACTCATAAGAACCTCGTGTGCAACTTAAAACAAGTCAAGTGCCCTGAAATGATGAAGGATGGTGTTGTGAAAGGTAAAGATGAGGTTGTTCTAACTGTTCCCCCGTTCTTCCATATTTACGGTTTCAACGGTATACTGAACTACAATCTTAGTCTTGGTTACCACATCGTTACTATGCCTAAATTTACTCCGGACGATTACATCAAGTGCTTAGTGGAACACGAACCGACCACACTGTTCGTGGTGCCATCGCTGTTGGCGTTCCTCGCGACTCATCCACTGGTGAAGAAGAGTCACCTTCAGTCCATTAAGATTATTATGGTCGGAGCGGCTCCCACCACAGACGCCATGATGGAGAAATTCTTGCTTAAGTGCGAGAAAACTAAAGATGATATCCGACTTTTGCAAGGGTACGGAATGACAGAGAGTTCTCCCGTGACTCTATTGACTCCGTTCGACTACCCTTACGGTAAGGTCGGCTCGGTCGGTCAACTAGTCCCCAGCACTCAGGCTAAAATCGTATCGCTGACCGATGGCGAAATGTTGGGGTCTCACAAGTCTGGAGAGTTATGGCTCAAAGGGCCTCAGGTGATGAAAGGCTATTGGAACAATGAAGCAGCGACCAAAGAGACTGTGGACAGCGACGGGTGGCTACACACCGGCGACGTTGCTTATTATGATGAGGATTATTATTTCTACATTGTTGACCGCACGAAAGAACTCATAAAAGTCAAAGGCAATCAGGTGTCACCAACTGAAGTGGAAAATGTTATATTGGAACGGCCCGAAGTAGCAGATGTGGCAGTCGTTGGCATTCCAGACCCTCTTGCTGGAGAACTACCTAAAGCCTTTGTAGTCTTGAAATCAAATCACAAACTAACAGAGAAAGAGGTTTATGATACAGTAGCACAAAAACTCACAAAGTACAAGCATCTCGAGGGTGGTGTAGTGTTTATGGATGCCATCCCAAGGAATGCTGCCGGAAAAATAATGAGAAACGAACTAAAAGTTTTAGGAGGAAAATCGAAACATTAA
Protein
MKYYTSQIDAQKNEEPIENLKNKTTLVTGKKINVKLKTSELKRLLGLAEPEKWTLTGAIGFLIVSSSVTMAVPFSLGKVLDIIYNSTSDLAAAREKLDALCLMLCGVFLIGGLCNFGRVYLMSISGQRMTQALRKQVYGAILKQETAWFGRTSTGELVNRLSADTQLVGRNLSQNVSDGLRSLFMVGAGTGMMFYMSPSLALIGLCVVPPVSMLAAIYGRFVRGITRELQDSLAETSELAEEKISNIKTVKAFSKEPKECESYAKRIENLLNLAYKESLAVGSFYGLTGLAGNTIIILVLYYGGGMVATEQLTVGNLTSFLLYAAYVGISIGGLSSFYTELNKGMGAATRLWEIIDREPKIPVTGGLIPKEKPNGEIILEKVHFSYNDAPLIRGLDLHLRPGKSVALVGRSGCGKSTIASLILRLYDPDKGRVLLDGVDIRELDPMWLRSLIGYVGQEPVLFSGTVKENILYGALDENEEVQNEKDEPAWMVAARTAHLHELVTGTRQGWDREVGAGGSQLSGGQKQRVAIARAIVKNPKILILDEATSALDTYSEYLVDKALKEISKSRTVLTIAHRLSTIQSADEVAVLENGVVVETGPYAELVAKPDGLFRELITHQTFTAKSRDKKDMQKSVLRRDFATRTATVSDSKCLSSPWGEIQIGKETLTDHVFGEVESWADKPSVTCGSSGRSYDYGMMRMMVERCANALVGHLKMKPGEKIGLILPNIPEFVVLIHGAMKAGLVVTFANPLYTAVEIQRQFNDCGVKAIATIEMFMPVAQEVSKSLKDYKGTIWVGGEDDKAKGIHGLRSLLMADHKADLPTLNCDDVCLIPYSSGTTGMPKGVMLTHKNLVCNLKQVKCPEMMKDGVVKGKDEVVLTVPPFFHIYGFNGILNYNLSLGYHIVTMPKFTPDDYIKCLVEHEPTTLFVVPSLLAFLATHPLVKKSHLQSIKIIMVGAAPTTDAMMEKFLLKCEKTKDDIRLLQGYGMTESSPVTLLTPFDYPYGKVGSVGQLVPSTQAKIVSLTDGEMLGSHKSGELWLKGPQVMKGYWNNEAATKETVDSDGWLHTGDVAYYDEDYYFYIVDRTKELIKVKGNQVSPTEVENVILERPEVADVAVVGIPDPLAGELPKAFVVLKSNHKLTEKEVYDTVAQKLTKYKHLEGGVVFMDAIPRNAAGKIMRNELKVLGGKSKH
Summary
Uniprot
Pubmed
EMBL
Proteomes
Interpro
Gene 3D
ProteinModelPortal
PDB
4AYX
E-value=8.27415e-168,
Score=1520
Ontologies
GO
PANTHER
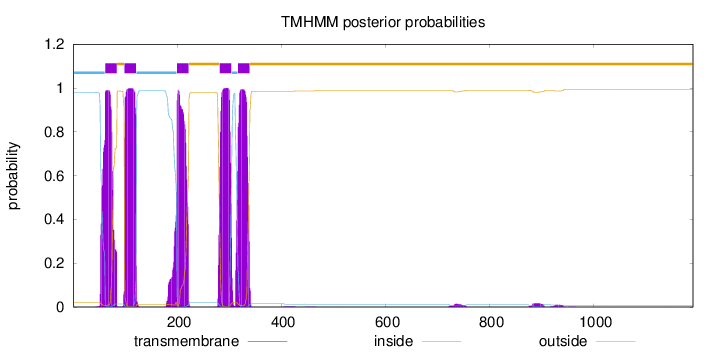
Topology
Length:
1191
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
113.690840000001
Exp number, first 60 AAs:
4.69361
Total prob of N-in:
0.98014
inside
1 - 61
TMhelix
62 - 84
outside
85 - 98
TMhelix
99 - 121
inside
122 - 199
TMhelix
200 - 222
outside
223 - 281
TMhelix
282 - 304
inside
305 - 316
TMhelix
317 - 339
outside
340 - 1191
Population Genetic Test Statistics
Pi
238.786696
Theta
176.408673
Tajima's D
0.990925
CLR
0.222066
CSRT
0.655617219139043
Interpretation
Uncertain
