Gene
KWMTBOMO10997 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008521
Annotation
PREDICTED:_xaa-Pro_dipeptidase_isoform_X2_[Bombyx_mori]
Full name
Xaa-Pro dipeptidase
Alternative Name
Imidodipeptidase
Peptidase D
Proline dipeptidase
Peptidase D
Proline dipeptidase
Location in the cell
Cytoplasmic Reliability : 1.583
Sequence
CDS
ATGGCAGCGACGTGGTCTATGGGCCCTGGCACTCTTGAAGTGCCGTTATCTTTGTTCGCAACAAACCGACGAAGATTAGCAAATAAACTCAAAAGTGGACAAATAGTGGTTTTGCAAGGAGGTGAAGATGTGAACCACTATGATACTGATGTACAATATGTATTTAGACAGGAAGCCTATTTTACATGGGTTTGTGGCGTTAGAGAACCCGGTTGTTATTTTGCCCTAGATGTTAGTACAGGCAAGTCTTACCTCTTTGTGCCCAGACTTCCTGAGGAGTATGAAGTCTGGATGGGCAAACTCCATGCCTGTAGTGACTTCAAAAACATATATGCAGTTGATGAAGTCTATTATGTTGATGAGATAAAAGATGTACTCAAAAGTTTGATGCCGGAAACATTGCTGACATTGTCTGGCCCAAACACAGACAGTGGCTTGACGGCCCGAGAAGCCATTTTTAATGGAATTGATGAGTTCAATGTGGATAATGAATCCTTATTTCCCATTATCGCAGAATTGTAA
Protein
MAATWSMGPGTLEVPLSLFATNRRRLANKLKSGQIVVLQGGEDVNHYDTDVQYVFRQEAYFTWVCGVREPGCYFALDVSTGKSYLFVPRLPEEYEVWMGKLHACSDFKNIYAVDEVYYVDEIKDVLKSLMPETLLTLSGPNTDSGLTAREAIFNGIDEFNVDNESLFPIIAEL
Summary
Catalytic Activity
Hydrolysis of Xaa-|-Pro dipeptides. Also acts on aminoacyl-hydroxyproline analogs. No action on Pro-|-Pro.
Cofactor
Mn(2+)
Subunit
Homodimer.
Similarity
Belongs to the peptidase M24B family.
Belongs to the peptidase M24B family. Eukaryotic-type prolidase subfamily.
Belongs to the peptidase M24B family. Eukaryotic-type prolidase subfamily.
Keywords
3D-structure
Acetylation
Alternative splicing
Collagen degradation
Complete proteome
Dipeptidase
Direct protein sequencing
Disease mutation
Hydrolase
Manganese
Metal-binding
Metalloprotease
Phosphoprotein
Polymorphism
Protease
Reference proteome
Feature
chain Xaa-Pro dipeptidase
splice variant In isoform 3.
sequence variant In PD; dbSNP:rs121917722.
splice variant In isoform 3.
sequence variant In PD; dbSNP:rs121917722.
Uniprot
H9JG74
A0A2A4JXP1
A0A2A4JVW9
A0A194PIF1
A0A194RRJ5
A0A2H1VH66
+ More
A0A212FGA3 A0A1I9WLD4 V5H133 A0A0L7LBM2 A0A2A3EBH4 T1DP25 A0A2I0M7X1 A0A2M4CTM5 J3JUK6 A0A1B6MCZ8 A0A2J8QGT9 A0A2Y9I4E4 A0A2M4CTQ3 A0A2M4CTP3 A0A091GZU2 A0A2M4AEE3 Q16XF1 A0A1S4FKR6 A0A0P6JU66 A0A2M4AE57 G5BXM2 A0A139WNR5 A0A151N708 A0A1L8DQK8 A0A1L8DQT9 A0A2J8RL00 D2I4G0 G1LCC7 A0A091DD12 U4UG94 N6TDH5 L5JMG2 A0A093HVV7 A0A093D0S0 K7FLR7 A0A023EVA5 E2BCC7 A0A1U7QVI0 A0A1V4K0N9 A0A091NIJ0 Q5ZKL3 A0A091GJ56 I3MA31 A0A091V531 A0A087R8U6 B1PMA7 A0A091WT01 A0A1B0FV12 A0A026W6M7 A0A1U7RLW2 A0A0A0ABK4 H0ZGS5 A0A158NBP2 U5EJJ7 G1N2P4 A0A094L6I4 A0A2R9A450 A0A023ESZ2 P12955-2 A0A2J8QH08 A0A1Q3FH32 A0A087V907 A0A093II33 A0A2I3RB40 A0A218UIL1 A0A1Q3FHW7 A0A140VJR2 P12955 J3K000 A0A2R8ZZT3 A0A2R9A0Z7 A0A2K6DUR6 A0A2R9A5F1 G2HFR0 A0A2I3S7Z0 A0A2R8ZZV1 U3ILP7 A0A034VC08 A0A2I3TWC3 A0A2I3RNM8 A0A2R8ZZU5 A0A3Q7UV47 R0L601 A0A093QN59 A0A2K6DUN1 A0A2Y9PFW1 A0A2K6DUQ2 A0A2K6DUP6 M3W8R0 A0A2K6DUQ3 A0A2I2V3Z9 A0A1I8G1B6 A0A3Q7VQ27 A0A091JPD1
A0A212FGA3 A0A1I9WLD4 V5H133 A0A0L7LBM2 A0A2A3EBH4 T1DP25 A0A2I0M7X1 A0A2M4CTM5 J3JUK6 A0A1B6MCZ8 A0A2J8QGT9 A0A2Y9I4E4 A0A2M4CTQ3 A0A2M4CTP3 A0A091GZU2 A0A2M4AEE3 Q16XF1 A0A1S4FKR6 A0A0P6JU66 A0A2M4AE57 G5BXM2 A0A139WNR5 A0A151N708 A0A1L8DQK8 A0A1L8DQT9 A0A2J8RL00 D2I4G0 G1LCC7 A0A091DD12 U4UG94 N6TDH5 L5JMG2 A0A093HVV7 A0A093D0S0 K7FLR7 A0A023EVA5 E2BCC7 A0A1U7QVI0 A0A1V4K0N9 A0A091NIJ0 Q5ZKL3 A0A091GJ56 I3MA31 A0A091V531 A0A087R8U6 B1PMA7 A0A091WT01 A0A1B0FV12 A0A026W6M7 A0A1U7RLW2 A0A0A0ABK4 H0ZGS5 A0A158NBP2 U5EJJ7 G1N2P4 A0A094L6I4 A0A2R9A450 A0A023ESZ2 P12955-2 A0A2J8QH08 A0A1Q3FH32 A0A087V907 A0A093II33 A0A2I3RB40 A0A218UIL1 A0A1Q3FHW7 A0A140VJR2 P12955 J3K000 A0A2R8ZZT3 A0A2R9A0Z7 A0A2K6DUR6 A0A2R9A5F1 G2HFR0 A0A2I3S7Z0 A0A2R8ZZV1 U3ILP7 A0A034VC08 A0A2I3TWC3 A0A2I3RNM8 A0A2R8ZZU5 A0A3Q7UV47 R0L601 A0A093QN59 A0A2K6DUN1 A0A2Y9PFW1 A0A2K6DUQ2 A0A2K6DUP6 M3W8R0 A0A2K6DUQ3 A0A2I2V3Z9 A0A1I8G1B6 A0A3Q7VQ27 A0A091JPD1
EC Number
3.4.13.9
Pubmed
19121390
26354079
22118469
27538518
26227816
23371554
+ More
22516182 17510324 21993625 18362917 19820115 22293439 20010809 23537049 23258410 17381049 24945155 20798317 15642098 24508170 20360741 21347285 20838655 22722832 2925654 14702039 15057824 15489334 11840567 19413330 21269460 22223895 24275569 2365824 8198124 8900231 12384772 16136131 21484476 23749191 25348373 17975172 26392545
22516182 17510324 21993625 18362917 19820115 22293439 20010809 23537049 23258410 17381049 24945155 20798317 15642098 24508170 20360741 21347285 20838655 22722832 2925654 14702039 15057824 15489334 11840567 19413330 21269460 22223895 24275569 2365824 8198124 8900231 12384772 16136131 21484476 23749191 25348373 17975172 26392545
EMBL
BABH01002123
BABH01002124
BABH01002125
NWSH01000474
PCG76152.1
PCG76151.1
+ More
KQ459604 KPI92504.1 KQ459765 KPJ20019.1 ODYU01002540 SOQ40166.1 AGBW02008690 OWR52762.1 KU932318 APA33954.1 GALX01001918 JAB66548.1 JTDY01001819 KOB72794.1 KZ288311 PBC28536.1 GAMD01002330 JAA99260.1 AKCR02000031 PKK25775.1 GGFL01004528 MBW68706.1 BT126921 AEE61883.1 GEBQ01006169 JAT33808.1 NBAG03000037 PNI95505.1 GGFL01004554 MBW68732.1 GGFL01004529 MBW68707.1 KL515729 KFO87935.1 GGFK01005799 MBW39120.1 CH477540 EAT39285.1 GEBF01005959 JAN97673.1 GGFK01005754 MBW39075.1 JH172334 EHB14033.1 KQ971307 KYB29640.1 AKHW03003905 KYO32612.1 GFDF01005341 JAV08743.1 GFDF01005340 JAV08744.1 NDHI03003675 PNJ09202.1 GL194465 EFB25831.1 ACTA01068868 KN122516 KFO30034.1 KB632309 ERL92052.1 APGK01042262 APGK01042263 KB741002 ENN75793.1 KB031158 ELK00530.1 KL206526 KFV83137.1 KL470553 KFV18704.1 AGCU01024232 AGCU01024233 AGCU01024234 AGCU01024235 AGCU01024236 AGCU01024237 AGCU01024238 AGCU01024239 AGCU01024240 AGCU01024241 GAPW01001184 JAC12414.1 GL447257 EFN86663.1 LSYS01005191 OPJ77943.1 KL383988 KFP89281.1 AJ720071 CAG31730.1 KL448230 KFO81341.1 AGTP01062927 AGTP01062928 AGTP01062929 KL410586 KFQ97482.1 KL226217 KFM09900.1 EU477530 ACA48584.1 KK734143 KFR04606.1 AJWK01006525 AJWK01006526 AJWK01006527 AJWK01006528 AJWK01006529 KK107453 EZA50679.1 KL871093 KGL90360.1 ABQF01043613 ABQF01043614 ABQF01043615 ABQF01043616 ABQF01043617 ABQF01043618 ABQF01043619 ABQF01043620 ABQF01043621 ABQF01043622 ADTU01011261 ADTU01011262 ADTU01011263 ADTU01011264 GANO01002212 JAB57659.1 KL360427 KFZ65087.1 AJFE02054631 AJFE02054632 AJFE02054633 AJFE02054634 AJFE02054635 AJFE02054636 AJFE02054637 AJFE02054638 AJFE02054639 GAPW01001185 JAC12413.1 J04605 BT006692 AK290756 AK290781 AK291561 AK293126 AK294619 AC008744 AC010485 BC004305 BC015027 BC028295 PNI95501.1 GFDL01008165 JAV26880.1 KL484381 KFO09099.1 KK594629 KFW02380.1 AACZ04019762 AACZ04019763 MUZQ01000278 OWK53627.1 GFDL01007898 JAV27147.1 HM005451 AEE61049.1 CR541669 CAG46470.1 AK305574 GABC01008843 GABF01009690 GABD01006559 GABE01010860 BAK62568.1 JAA02495.1 JAA12455.1 JAA26541.1 JAA33879.1 PNI95500.1 ADON01076040 ADON01076041 ADON01076042 ADON01076043 GAKP01019316 GAKP01019315 JAC39637.1 KB743139 EOB01044.1 KL672275 KFW85417.1 AANG04004361 NIVC01003675 PAA50308.1 KK501247 KFP13531.1
KQ459604 KPI92504.1 KQ459765 KPJ20019.1 ODYU01002540 SOQ40166.1 AGBW02008690 OWR52762.1 KU932318 APA33954.1 GALX01001918 JAB66548.1 JTDY01001819 KOB72794.1 KZ288311 PBC28536.1 GAMD01002330 JAA99260.1 AKCR02000031 PKK25775.1 GGFL01004528 MBW68706.1 BT126921 AEE61883.1 GEBQ01006169 JAT33808.1 NBAG03000037 PNI95505.1 GGFL01004554 MBW68732.1 GGFL01004529 MBW68707.1 KL515729 KFO87935.1 GGFK01005799 MBW39120.1 CH477540 EAT39285.1 GEBF01005959 JAN97673.1 GGFK01005754 MBW39075.1 JH172334 EHB14033.1 KQ971307 KYB29640.1 AKHW03003905 KYO32612.1 GFDF01005341 JAV08743.1 GFDF01005340 JAV08744.1 NDHI03003675 PNJ09202.1 GL194465 EFB25831.1 ACTA01068868 KN122516 KFO30034.1 KB632309 ERL92052.1 APGK01042262 APGK01042263 KB741002 ENN75793.1 KB031158 ELK00530.1 KL206526 KFV83137.1 KL470553 KFV18704.1 AGCU01024232 AGCU01024233 AGCU01024234 AGCU01024235 AGCU01024236 AGCU01024237 AGCU01024238 AGCU01024239 AGCU01024240 AGCU01024241 GAPW01001184 JAC12414.1 GL447257 EFN86663.1 LSYS01005191 OPJ77943.1 KL383988 KFP89281.1 AJ720071 CAG31730.1 KL448230 KFO81341.1 AGTP01062927 AGTP01062928 AGTP01062929 KL410586 KFQ97482.1 KL226217 KFM09900.1 EU477530 ACA48584.1 KK734143 KFR04606.1 AJWK01006525 AJWK01006526 AJWK01006527 AJWK01006528 AJWK01006529 KK107453 EZA50679.1 KL871093 KGL90360.1 ABQF01043613 ABQF01043614 ABQF01043615 ABQF01043616 ABQF01043617 ABQF01043618 ABQF01043619 ABQF01043620 ABQF01043621 ABQF01043622 ADTU01011261 ADTU01011262 ADTU01011263 ADTU01011264 GANO01002212 JAB57659.1 KL360427 KFZ65087.1 AJFE02054631 AJFE02054632 AJFE02054633 AJFE02054634 AJFE02054635 AJFE02054636 AJFE02054637 AJFE02054638 AJFE02054639 GAPW01001185 JAC12413.1 J04605 BT006692 AK290756 AK290781 AK291561 AK293126 AK294619 AC008744 AC010485 BC004305 BC015027 BC028295 PNI95501.1 GFDL01008165 JAV26880.1 KL484381 KFO09099.1 KK594629 KFW02380.1 AACZ04019762 AACZ04019763 MUZQ01000278 OWK53627.1 GFDL01007898 JAV27147.1 HM005451 AEE61049.1 CR541669 CAG46470.1 AK305574 GABC01008843 GABF01009690 GABD01006559 GABE01010860 BAK62568.1 JAA02495.1 JAA12455.1 JAA26541.1 JAA33879.1 PNI95500.1 ADON01076040 ADON01076041 ADON01076042 ADON01076043 GAKP01019316 GAKP01019315 JAC39637.1 KB743139 EOB01044.1 KL672275 KFW85417.1 AANG04004361 NIVC01003675 PAA50308.1 KK501247 KFP13531.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000037510
+ More
UP000242457 UP000053872 UP000248481 UP000008820 UP000006813 UP000007266 UP000050525 UP000008912 UP000028990 UP000030742 UP000019118 UP000010552 UP000053584 UP000007267 UP000008237 UP000189706 UP000190648 UP000053760 UP000005215 UP000053283 UP000053286 UP000053605 UP000092461 UP000053097 UP000189705 UP000053858 UP000007754 UP000005205 UP000001645 UP000240080 UP000005640 UP000002277 UP000197619 UP000233120 UP000016666 UP000286642 UP000053258 UP000248483 UP000011712 UP000095280 UP000215902 UP000053119
UP000242457 UP000053872 UP000248481 UP000008820 UP000006813 UP000007266 UP000050525 UP000008912 UP000028990 UP000030742 UP000019118 UP000010552 UP000053584 UP000007267 UP000008237 UP000189706 UP000190648 UP000053760 UP000005215 UP000053283 UP000053286 UP000053605 UP000092461 UP000053097 UP000189705 UP000053858 UP000007754 UP000005205 UP000001645 UP000240080 UP000005640 UP000002277 UP000197619 UP000233120 UP000016666 UP000286642 UP000053258 UP000248483 UP000011712 UP000095280 UP000215902 UP000053119
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JG74
A0A2A4JXP1
A0A2A4JVW9
A0A194PIF1
A0A194RRJ5
A0A2H1VH66
+ More
A0A212FGA3 A0A1I9WLD4 V5H133 A0A0L7LBM2 A0A2A3EBH4 T1DP25 A0A2I0M7X1 A0A2M4CTM5 J3JUK6 A0A1B6MCZ8 A0A2J8QGT9 A0A2Y9I4E4 A0A2M4CTQ3 A0A2M4CTP3 A0A091GZU2 A0A2M4AEE3 Q16XF1 A0A1S4FKR6 A0A0P6JU66 A0A2M4AE57 G5BXM2 A0A139WNR5 A0A151N708 A0A1L8DQK8 A0A1L8DQT9 A0A2J8RL00 D2I4G0 G1LCC7 A0A091DD12 U4UG94 N6TDH5 L5JMG2 A0A093HVV7 A0A093D0S0 K7FLR7 A0A023EVA5 E2BCC7 A0A1U7QVI0 A0A1V4K0N9 A0A091NIJ0 Q5ZKL3 A0A091GJ56 I3MA31 A0A091V531 A0A087R8U6 B1PMA7 A0A091WT01 A0A1B0FV12 A0A026W6M7 A0A1U7RLW2 A0A0A0ABK4 H0ZGS5 A0A158NBP2 U5EJJ7 G1N2P4 A0A094L6I4 A0A2R9A450 A0A023ESZ2 P12955-2 A0A2J8QH08 A0A1Q3FH32 A0A087V907 A0A093II33 A0A2I3RB40 A0A218UIL1 A0A1Q3FHW7 A0A140VJR2 P12955 J3K000 A0A2R8ZZT3 A0A2R9A0Z7 A0A2K6DUR6 A0A2R9A5F1 G2HFR0 A0A2I3S7Z0 A0A2R8ZZV1 U3ILP7 A0A034VC08 A0A2I3TWC3 A0A2I3RNM8 A0A2R8ZZU5 A0A3Q7UV47 R0L601 A0A093QN59 A0A2K6DUN1 A0A2Y9PFW1 A0A2K6DUQ2 A0A2K6DUP6 M3W8R0 A0A2K6DUQ3 A0A2I2V3Z9 A0A1I8G1B6 A0A3Q7VQ27 A0A091JPD1
A0A212FGA3 A0A1I9WLD4 V5H133 A0A0L7LBM2 A0A2A3EBH4 T1DP25 A0A2I0M7X1 A0A2M4CTM5 J3JUK6 A0A1B6MCZ8 A0A2J8QGT9 A0A2Y9I4E4 A0A2M4CTQ3 A0A2M4CTP3 A0A091GZU2 A0A2M4AEE3 Q16XF1 A0A1S4FKR6 A0A0P6JU66 A0A2M4AE57 G5BXM2 A0A139WNR5 A0A151N708 A0A1L8DQK8 A0A1L8DQT9 A0A2J8RL00 D2I4G0 G1LCC7 A0A091DD12 U4UG94 N6TDH5 L5JMG2 A0A093HVV7 A0A093D0S0 K7FLR7 A0A023EVA5 E2BCC7 A0A1U7QVI0 A0A1V4K0N9 A0A091NIJ0 Q5ZKL3 A0A091GJ56 I3MA31 A0A091V531 A0A087R8U6 B1PMA7 A0A091WT01 A0A1B0FV12 A0A026W6M7 A0A1U7RLW2 A0A0A0ABK4 H0ZGS5 A0A158NBP2 U5EJJ7 G1N2P4 A0A094L6I4 A0A2R9A450 A0A023ESZ2 P12955-2 A0A2J8QH08 A0A1Q3FH32 A0A087V907 A0A093II33 A0A2I3RB40 A0A218UIL1 A0A1Q3FHW7 A0A140VJR2 P12955 J3K000 A0A2R8ZZT3 A0A2R9A0Z7 A0A2K6DUR6 A0A2R9A5F1 G2HFR0 A0A2I3S7Z0 A0A2R8ZZV1 U3ILP7 A0A034VC08 A0A2I3TWC3 A0A2I3RNM8 A0A2R8ZZU5 A0A3Q7UV47 R0L601 A0A093QN59 A0A2K6DUN1 A0A2Y9PFW1 A0A2K6DUQ2 A0A2K6DUP6 M3W8R0 A0A2K6DUQ3 A0A2I2V3Z9 A0A1I8G1B6 A0A3Q7VQ27 A0A091JPD1
PDB
5MC5
E-value=8.11885e-50,
Score=493
Ontologies
GO
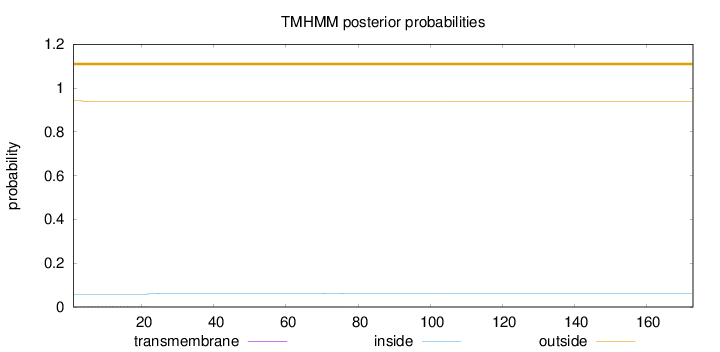
Topology
Length:
173
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.06532
Exp number, first 60 AAs:
0.06241
Total prob of N-in:
0.05833
outside
1 - 173
Population Genetic Test Statistics
Pi
262.340002
Theta
208.046834
Tajima's D
0.410349
CLR
0.001955
CSRT
0.488025598720064
Interpretation
Uncertain
