Gene
KWMTBOMO10991
Pre Gene Modal
BGIBMGA008518
Annotation
PREDICTED:_histone-lysine_N-methyltransferase_setd3-like_[Amyelois_transitella]
Full name
Actin-histidine N-methyltransferase
Alternative Name
SET domain-containing protein 3
Location in the cell
Cytoplasmic Reliability : 1.071 Mitochondrial Reliability : 1.668 Nuclear Reliability : 1.516
Sequence
CDS
ATGGGACGAAAATCGCAATCAAAACAAAACCAAAAGAAATCCACTGGAAAGGAAAATAATAAATTATACCTACAGAAAAGAAAAGAATTAGCTTTTCTTGTTGATAGAGTGCTAAGACTTACCAGTATATTTCAAGCGACTGGTAACGTAAGCAAAAGTTGGGAACATCATTTAGAAATCGAGAAATTAGTGAATGAAATTACTAACATAGAAGGAACGCAGTTAAGAAATACTCGAACTCTTCGGCAAACTCACATTGATAAATATTTGCAGTGGCTAAGAGAAAACGGTGTCCAATTCGATGGGATTGAAATAGCGGAATTCAAAGGATATGATCTTGGTCTTAAAGCTTTGAAAGATTTCACTGAAGGATCTCTGATTTTAACCATACCATCCAAAGTTATGATTACAGAGAAAAATGCCAAAGAATCTGAACTATCAGCATTTATTGCTGTCGATCCTCTTTTACAGAATATGCCAAATATTACTTTAGCCCTATTCCTATTACTTGAAAATAATAACCCAGATTCATTCTGGAAGCCGTACATTGAGGTTTTGCCTGAAAAATATTCTACCATTTTATACTACACATCTGAAGAACTTGAACTATTGAAACCATCGCCAGTGTTTGAATCATCACTAAAACTTTACAGAAGTATAGCCAGACAGTATGCATACTTTTATAAAAAGATTCACACACTCGATATACCAGTGCTGAAGAAACTTCAAGAAGTTTTCACTTTTGATAGTTACAGGTGGGCAGTCTCAACCGTGATGACAAGGCAAAACAATATAAGACTTTCGGAAGCTGACGTCACAGCGTTCATACCGCTATGGGATATGTGCAACCATGAACACGGCGAGATAAGAACGGACTACAACAACGAACTAAATAGAGGCGAATGCTACGCGCTCCGCGATTTTAAAACCGACGAACAAATCTTCATATTCTACGGAGCTCGTCCTAACGCAGACCTGTTTCTACACAATGGTTTTGTTTACCCGAATAATCAGCACGACAGCCTATCTGTAACACTCGGCATTAGCTCGGGCGATCCTCTGCGAGAGATAAAAATGTCTCTGCTCACGAAGCTAGGGCTGGGCTGCGTCACGCACTACTCTCTGTTCAAACGACAGAATCCAATCGGACCAGAACTATTGGCGTTTATAAGGATCTTCAATATGAATCAGGAGGAACTAACGAAATGGTCCAGTCAAAGTCTTCCAAGCGACCTTGTGTCGTCGGAGCCGAGCAGCGCTGAGGCGGTAGGCGCCGACATCGACCGTCGAGCCCACACGTATCTACTGACGCGGTGCAAACTGCTACTAGCTGCATACAAGAAAAACGACAACGAAAATCGCACAGAAACCGATCATGTTTCCAACGTGAAACTCTTGAAACAATGCGAAGTACAAATACTGGAAGCTACTGTTACGTACCTGGAAGAAACTATACGAACACTACCCACCGCGGTTAAACAATAA
Protein
MGRKSQSKQNQKKSTGKENNKLYLQKRKELAFLVDRVLRLTSIFQATGNVSKSWEHHLEIEKLVNEITNIEGTQLRNTRTLRQTHIDKYLQWLRENGVQFDGIEIAEFKGYDLGLKALKDFTEGSLILTIPSKVMITEKNAKESELSAFIAVDPLLQNMPNITLALFLLLENNNPDSFWKPYIEVLPEKYSTILYYTSEELELLKPSPVFESSLKLYRSIARQYAYFYKKIHTLDIPVLKKLQEVFTFDSYRWAVSTVMTRQNNIRLSEADVTAFIPLWDMCNHEHGEIRTDYNNELNRGECYALRDFKTDEQIFIFYGARPNADLFLHNGFVYPNNQHDSLSVTLGISSGDPLREIKMSLLTKLGLGCVTHYSLFKRQNPIGPELLAFIRIFNMNQEELTKWSSQSLPSDLVSSEPSSAEAVGADIDRRAHTYLLTRCKLLLAAYKKNDNENRTETDHVSNVKLLKQCEVQILEATVTYLEETIRTLPTAVKQ
Summary
Description
Protein-histidine N-methyltransferase that specifically mediates methylation of actin at 'His-73'. Does not have protein-lysine N-methyltransferase activity and probably only catalyzes histidine methylation of actin.
Catalytic Activity
L-histidyl-[protein] + S-adenosyl-L-methionine = H(+) + N(tele)-methyl-L-histidyl-[protein] + S-adenosyl-L-homocysteine
Similarity
Belongs to the class V-like SAM-binding methyltransferase superfamily. Histone-lysine methyltransferase family. SETD3 subfamily.
Keywords
Actin-binding
Complete proteome
Cytoplasm
Methyltransferase
Reference proteome
S-adenosyl-L-methionine
Transferase
Feature
chain Actin-histidine N-methyltransferase
Uniprot
A0A2A4JWE5
S4PTC4
A0A212EYG4
A0A2J7RSA4
A0A2J7RS85
A0A1Y1M7T9
+ More
A0A1B6EQS2 A0A1W4XMC6 A0A1B6DPA2 A0A1B6K1K7 A0A067R8M1 A0A1S3IK48 A0A1S3KBJ0 A0A3R7MC43 A0A224XBD5 A0A023F1B6 A0A210QXD5 A0A151IHD2 A0A151WMU9 A0A195F9F4 A0A195EDE1 A0A0P4WKD1 A0A2C9KUH6 A0A0P4W6C3 A0A0B7BBI2 A0A2T7NXF7 A0A3L8DG84 A0A0L8FSI6 A0A026X153 V4AGI2 X1WIN4 H2S936 C3Z3T7 A0A2H8TI53 E0VEE0 A0A3Q1IHH0 A0A3P8X932 A0A3P8XDF4 A0A3Q3MK49 A0A2I4BHU5 A0A3Q1IK56 A0A3Q3MZG9 A0A2U9CKC8 A0A091WA52 R7UY49 Q5ZML9 A0A1E1XT20 A0A3Q3MK07 A0A0C9PZ01 A0A1L1RV17 U3IE49 A0A091V826 H3CS80 A0A093FVS8 A0A3Q2WB75 A0A2S2NSK9 A0A1D5PBS7 G1NKN9 A0A0A9WTQ0 A0A3B1K7Y5 A0A091HKK3 U6DBW1 A0A093HG15 A0A093NN10 A0A3Q3JEP2 A0A3B3S7Q8 A0A3B4FRC4 A0A087R567 A0A0A0AW32 A0A091RLU2 A0A131YZ62 W5L904 Q7SXS7 A0A2I0MP26 A0A3P8NSZ4 A0A151P9L2 A0A224Z030 A0A0Q3M4I5 A0A2A3E4F5 A0A1L8F9M7 A0A2I0TQF3 A0A3P9CLY9 R0JSN7 A0A3Q2UU46 A0A2Y9JJ92 A0A0P5P6X1 A0A099ZM06 A0A091FYZ1 A0A0N8CGI8 A0A3Q3FJ49 A0A1S2ZF79 A0A1V4KAW4 A0A2Y9JJE3 A0A3B4EAC8
A0A1B6EQS2 A0A1W4XMC6 A0A1B6DPA2 A0A1B6K1K7 A0A067R8M1 A0A1S3IK48 A0A1S3KBJ0 A0A3R7MC43 A0A224XBD5 A0A023F1B6 A0A210QXD5 A0A151IHD2 A0A151WMU9 A0A195F9F4 A0A195EDE1 A0A0P4WKD1 A0A2C9KUH6 A0A0P4W6C3 A0A0B7BBI2 A0A2T7NXF7 A0A3L8DG84 A0A0L8FSI6 A0A026X153 V4AGI2 X1WIN4 H2S936 C3Z3T7 A0A2H8TI53 E0VEE0 A0A3Q1IHH0 A0A3P8X932 A0A3P8XDF4 A0A3Q3MK49 A0A2I4BHU5 A0A3Q1IK56 A0A3Q3MZG9 A0A2U9CKC8 A0A091WA52 R7UY49 Q5ZML9 A0A1E1XT20 A0A3Q3MK07 A0A0C9PZ01 A0A1L1RV17 U3IE49 A0A091V826 H3CS80 A0A093FVS8 A0A3Q2WB75 A0A2S2NSK9 A0A1D5PBS7 G1NKN9 A0A0A9WTQ0 A0A3B1K7Y5 A0A091HKK3 U6DBW1 A0A093HG15 A0A093NN10 A0A3Q3JEP2 A0A3B3S7Q8 A0A3B4FRC4 A0A087R567 A0A0A0AW32 A0A091RLU2 A0A131YZ62 W5L904 Q7SXS7 A0A2I0MP26 A0A3P8NSZ4 A0A151P9L2 A0A224Z030 A0A0Q3M4I5 A0A2A3E4F5 A0A1L8F9M7 A0A2I0TQF3 A0A3P9CLY9 R0JSN7 A0A3Q2UU46 A0A2Y9JJ92 A0A0P5P6X1 A0A099ZM06 A0A091FYZ1 A0A0N8CGI8 A0A3Q3FJ49 A0A1S2ZF79 A0A1V4KAW4 A0A2Y9JJE3 A0A3B4EAC8
EC Number
2.1.1.85
Pubmed
EMBL
NWSH01000474
PCG76139.1
GAIX01012283
JAA80277.1
AGBW02011509
OWR46539.1
+ More
NEVH01000267 PNF43702.1 PNF43701.1 GEZM01038162 JAV81753.1 GECZ01029475 JAS40294.1 GEDC01009783 JAS27515.1 GECU01002382 JAT05325.1 KK852624 KDR19932.1 QCYY01001344 ROT78720.1 GFTR01006761 JAW09665.1 GBBI01003634 JAC15078.1 NEDP02001349 OWF53384.1 KQ977613 KYN01385.1 KQ982934 KYQ49199.1 KQ981727 KYN37011.1 KQ979074 KYN23111.1 GDRN01083261 GDRN01083258 JAI61700.1 GDRN01083260 GDRN01083259 JAI61702.1 HACG01042786 HACG01042788 CEK89651.1 CEK89653.1 PZQS01000008 PVD25870.1 QOIP01000008 RLU19316.1 KQ426865 KOF67661.1 KK107034 EZA62035.1 KB201847 ESO94275.1 ABLF02040907 GG666577 EEN52851.1 GFXV01002000 MBW13805.1 AAZO01001525 DS235090 EEB11746.1 CP026259 AWP16977.1 KK735116 KFR12537.1 AMQN01005800 KB296865 ELU11224.1 AJ719365 AADN02003714 AADN02003715 GFAA01001001 JAU02434.1 GBYB01006733 JAG76500.1 AADN05000303 AC183980 ADON01080962 ADON01080963 KL410770 KFQ99446.1 KL214832 KFV62030.1 GGMR01007393 MBY20012.1 GBHO01032490 JAG11114.1 KL217494 KFO95729.1 HAAF01008250 CCP80074.1 KL206356 KFV81588.1 KL224603 KFW61552.1 KL226108 KFM08621.1 KL873269 KGL98077.1 KK802075 KFQ29492.1 GEDV01004609 JAP83948.1 BC055261 BC155278 AKCR02000005 PKK31429.1 AKHW03000563 KYO45435.1 GFPF01008438 MAA19584.1 LMAW01002745 KQK77557.1 KZ288377 PBC26643.1 CM004480 OCT68299.1 KZ507896 PKU36039.1 KB743197 EOB00426.1 GDIQ01132792 JAL18934.1 KL894670 KGL81933.1 KL447555 KFO74883.1 GDIP01133943 LRGB01001348 JAL69771.1 KZS12928.1 LSYS01003973 OPJ81491.1
NEVH01000267 PNF43702.1 PNF43701.1 GEZM01038162 JAV81753.1 GECZ01029475 JAS40294.1 GEDC01009783 JAS27515.1 GECU01002382 JAT05325.1 KK852624 KDR19932.1 QCYY01001344 ROT78720.1 GFTR01006761 JAW09665.1 GBBI01003634 JAC15078.1 NEDP02001349 OWF53384.1 KQ977613 KYN01385.1 KQ982934 KYQ49199.1 KQ981727 KYN37011.1 KQ979074 KYN23111.1 GDRN01083261 GDRN01083258 JAI61700.1 GDRN01083260 GDRN01083259 JAI61702.1 HACG01042786 HACG01042788 CEK89651.1 CEK89653.1 PZQS01000008 PVD25870.1 QOIP01000008 RLU19316.1 KQ426865 KOF67661.1 KK107034 EZA62035.1 KB201847 ESO94275.1 ABLF02040907 GG666577 EEN52851.1 GFXV01002000 MBW13805.1 AAZO01001525 DS235090 EEB11746.1 CP026259 AWP16977.1 KK735116 KFR12537.1 AMQN01005800 KB296865 ELU11224.1 AJ719365 AADN02003714 AADN02003715 GFAA01001001 JAU02434.1 GBYB01006733 JAG76500.1 AADN05000303 AC183980 ADON01080962 ADON01080963 KL410770 KFQ99446.1 KL214832 KFV62030.1 GGMR01007393 MBY20012.1 GBHO01032490 JAG11114.1 KL217494 KFO95729.1 HAAF01008250 CCP80074.1 KL206356 KFV81588.1 KL224603 KFW61552.1 KL226108 KFM08621.1 KL873269 KGL98077.1 KK802075 KFQ29492.1 GEDV01004609 JAP83948.1 BC055261 BC155278 AKCR02000005 PKK31429.1 AKHW03000563 KYO45435.1 GFPF01008438 MAA19584.1 LMAW01002745 KQK77557.1 KZ288377 PBC26643.1 CM004480 OCT68299.1 KZ507896 PKU36039.1 KB743197 EOB00426.1 GDIQ01132792 JAL18934.1 KL894670 KGL81933.1 KL447555 KFO74883.1 GDIP01133943 LRGB01001348 JAL69771.1 KZS12928.1 LSYS01003973 OPJ81491.1
Proteomes
UP000218220
UP000007151
UP000235965
UP000192223
UP000027135
UP000085678
+ More
UP000283509 UP000242188 UP000078542 UP000075809 UP000078541 UP000078492 UP000076420 UP000245119 UP000279307 UP000053454 UP000053097 UP000030746 UP000007819 UP000005226 UP000001554 UP000009046 UP000265040 UP000265140 UP000261640 UP000192220 UP000246464 UP000053605 UP000014760 UP000000539 UP000016666 UP000053283 UP000007303 UP000053875 UP000264840 UP000001645 UP000018467 UP000054308 UP000053584 UP000054081 UP000261600 UP000261540 UP000261460 UP000053286 UP000053858 UP000000437 UP000053872 UP000265100 UP000050525 UP000051836 UP000242457 UP000186698 UP000265160 UP000248482 UP000053641 UP000053760 UP000076858 UP000261660 UP000079721 UP000190648 UP000261440
UP000283509 UP000242188 UP000078542 UP000075809 UP000078541 UP000078492 UP000076420 UP000245119 UP000279307 UP000053454 UP000053097 UP000030746 UP000007819 UP000005226 UP000001554 UP000009046 UP000265040 UP000265140 UP000261640 UP000192220 UP000246464 UP000053605 UP000014760 UP000000539 UP000016666 UP000053283 UP000007303 UP000053875 UP000264840 UP000001645 UP000018467 UP000054308 UP000053584 UP000054081 UP000261600 UP000261540 UP000261460 UP000053286 UP000053858 UP000000437 UP000053872 UP000265100 UP000050525 UP000051836 UP000242457 UP000186698 UP000265160 UP000248482 UP000053641 UP000053760 UP000076858 UP000261660 UP000079721 UP000190648 UP000261440
Interpro
SUPFAM
SSF81822
SSF81822
Gene 3D
ProteinModelPortal
A0A2A4JWE5
S4PTC4
A0A212EYG4
A0A2J7RSA4
A0A2J7RS85
A0A1Y1M7T9
+ More
A0A1B6EQS2 A0A1W4XMC6 A0A1B6DPA2 A0A1B6K1K7 A0A067R8M1 A0A1S3IK48 A0A1S3KBJ0 A0A3R7MC43 A0A224XBD5 A0A023F1B6 A0A210QXD5 A0A151IHD2 A0A151WMU9 A0A195F9F4 A0A195EDE1 A0A0P4WKD1 A0A2C9KUH6 A0A0P4W6C3 A0A0B7BBI2 A0A2T7NXF7 A0A3L8DG84 A0A0L8FSI6 A0A026X153 V4AGI2 X1WIN4 H2S936 C3Z3T7 A0A2H8TI53 E0VEE0 A0A3Q1IHH0 A0A3P8X932 A0A3P8XDF4 A0A3Q3MK49 A0A2I4BHU5 A0A3Q1IK56 A0A3Q3MZG9 A0A2U9CKC8 A0A091WA52 R7UY49 Q5ZML9 A0A1E1XT20 A0A3Q3MK07 A0A0C9PZ01 A0A1L1RV17 U3IE49 A0A091V826 H3CS80 A0A093FVS8 A0A3Q2WB75 A0A2S2NSK9 A0A1D5PBS7 G1NKN9 A0A0A9WTQ0 A0A3B1K7Y5 A0A091HKK3 U6DBW1 A0A093HG15 A0A093NN10 A0A3Q3JEP2 A0A3B3S7Q8 A0A3B4FRC4 A0A087R567 A0A0A0AW32 A0A091RLU2 A0A131YZ62 W5L904 Q7SXS7 A0A2I0MP26 A0A3P8NSZ4 A0A151P9L2 A0A224Z030 A0A0Q3M4I5 A0A2A3E4F5 A0A1L8F9M7 A0A2I0TQF3 A0A3P9CLY9 R0JSN7 A0A3Q2UU46 A0A2Y9JJ92 A0A0P5P6X1 A0A099ZM06 A0A091FYZ1 A0A0N8CGI8 A0A3Q3FJ49 A0A1S2ZF79 A0A1V4KAW4 A0A2Y9JJE3 A0A3B4EAC8
A0A1B6EQS2 A0A1W4XMC6 A0A1B6DPA2 A0A1B6K1K7 A0A067R8M1 A0A1S3IK48 A0A1S3KBJ0 A0A3R7MC43 A0A224XBD5 A0A023F1B6 A0A210QXD5 A0A151IHD2 A0A151WMU9 A0A195F9F4 A0A195EDE1 A0A0P4WKD1 A0A2C9KUH6 A0A0P4W6C3 A0A0B7BBI2 A0A2T7NXF7 A0A3L8DG84 A0A0L8FSI6 A0A026X153 V4AGI2 X1WIN4 H2S936 C3Z3T7 A0A2H8TI53 E0VEE0 A0A3Q1IHH0 A0A3P8X932 A0A3P8XDF4 A0A3Q3MK49 A0A2I4BHU5 A0A3Q1IK56 A0A3Q3MZG9 A0A2U9CKC8 A0A091WA52 R7UY49 Q5ZML9 A0A1E1XT20 A0A3Q3MK07 A0A0C9PZ01 A0A1L1RV17 U3IE49 A0A091V826 H3CS80 A0A093FVS8 A0A3Q2WB75 A0A2S2NSK9 A0A1D5PBS7 G1NKN9 A0A0A9WTQ0 A0A3B1K7Y5 A0A091HKK3 U6DBW1 A0A093HG15 A0A093NN10 A0A3Q3JEP2 A0A3B3S7Q8 A0A3B4FRC4 A0A087R567 A0A0A0AW32 A0A091RLU2 A0A131YZ62 W5L904 Q7SXS7 A0A2I0MP26 A0A3P8NSZ4 A0A151P9L2 A0A224Z030 A0A0Q3M4I5 A0A2A3E4F5 A0A1L8F9M7 A0A2I0TQF3 A0A3P9CLY9 R0JSN7 A0A3Q2UU46 A0A2Y9JJ92 A0A0P5P6X1 A0A099ZM06 A0A091FYZ1 A0A0N8CGI8 A0A3Q3FJ49 A0A1S2ZF79 A0A1V4KAW4 A0A2Y9JJE3 A0A3B4EAC8
PDB
3SMT
E-value=1.67614e-78,
Score=746
Ontologies
GO
GO:0046975
GO:0003713
GO:0008168
GO:0018026
GO:0018023
GO:0042800
GO:0000790
GO:0018027
GO:0045944
GO:0016279
GO:0030785
GO:0045893
GO:0018021
GO:0030047
GO:0005737
GO:0003779
GO:0018064
GO:0010452
GO:0051149
GO:0070472
GO:0001102
GO:0008283
GO:0001558
GO:0018022
GO:0005515
GO:0006357
GO:0051603
GO:0051028
GO:0005622
GO:0006810
GO:0003824
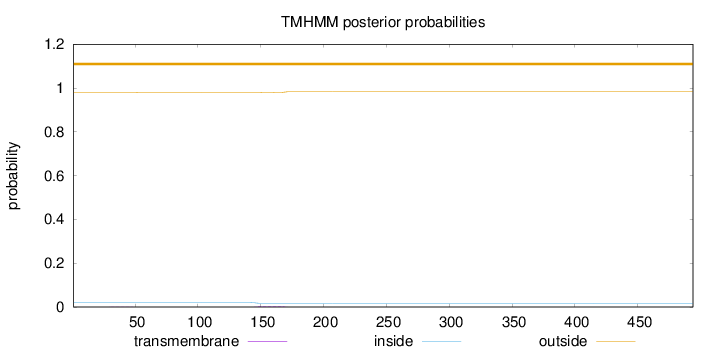
Topology
Subcellular location
Cytoplasm
Length:
494
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0677200000000001
Exp number, first 60 AAs:
0.00308
Total prob of N-in:
0.01993
outside
1 - 494
Population Genetic Test Statistics
Pi
237.960677
Theta
164.995032
Tajima's D
1.221462
CLR
0.212638
CSRT
0.724713764311784
Interpretation
Uncertain
