Gene
KWMTBOMO10986
Pre Gene Modal
BGIBMGA008513
Annotation
serine_protease_36_[Mamestra_configurata]
Location in the cell
Extracellular Reliability : 1.957 PlasmaMembrane Reliability : 2.405
Sequence
CDS
ATGAAGTTCATCATATTTCTCGGCTTAACAAATTTAGTCTGTGGTATAAATTTAAGTGAAACGTTTAGGAACAGCGCTTTCGGATATCATGCCAGGGTCGGAATTCCTTTAGCTGAAAGAATACAAAGAGATGAAAATAATTATTTCAATTCTAGAATTGTTGGAGGTGTACCAGCCGTCGCTGGACAATATCCTTTTAAAGCCGGACTAATTGGAGATATAATTGGATTTGATGGTGTTAGCGTGTGCGGTGGATCCCTTATCCATCCTTCAAAGGTTGCTACAGCTGCGCACTGCTGGAATGACGGTGAAAACCAAGCATGGCGTTTTACCGTCGTGCTTGGTTCTAGCTTATTGTTCAGCGGCGGAACGAGACTTGCTTCAAGCGTTGTTGTCGCTCATCCAGATTGGAACCCTAATTCGGTTCAGAATGATGTTGCTGTGATATATTTAACCTATGAAGTAGAAACATCAAGTATCATTACACCTATCGCTTTACCATCCGGTCCTGAACTGTTTGAAAATTTTAAAGGAGAAGCGGCAATTGCCATTGGATTTGGTATCACGAGTAATGATAACGGCATTTCATCTAATCAGTTCTTAAGTCATGTGCGCTTGACTGTAATTTCGAATAATATTTGCAATTTGGTGTTCCCGTTTATTTTGCAAGCATCCAACATCTGTACCAGTGGTTTTGGATTTATTGGTACTTGCGGTGGAGACTCCGGTGGTCCTCTTATTACTTACAGGAATAATGAGCCTATATTAATTGGCATTGCATCATTCGGTTCGGCCTTAGGTTGTGAAATCGGCCTCCCGTCTGCTTACTCCAGAGCAACATCATATGTGGACTTCTTCAACCAACACTTAAAACAATAA
Protein
MKFIIFLGLTNLVCGINLSETFRNSAFGYHARVGIPLAERIQRDENNYFNSRIVGGVPAVAGQYPFKAGLIGDIIGFDGVSVCGGSLIHPSKVATAAHCWNDGENQAWRFTVVLGSSLLFSGGTRLASSVVVAHPDWNPNSVQNDVAVIYLTYEVETSSIITPIALPSGPELFENFKGEAAIAIGFGITSNDNGISSNQFLSHVRLTVISNNICNLVFPFILQASNICTSGFGFIGTCGGDSGGPLITYRNNEPILIGIASFGSALGCEIGLPSAYSRATSYVDFFNQHLKQ
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
A0A1B1LTS0
E0W9N4
C9W8F6
A0A2A4K0B4
A0A2H1V6P6
A0A2H1VW13
+ More
A0A2A4K202 A0A2H1V6W6 A0A089QAL5 A0A0K8SR13 A0A2H1VX12 A0A0K8SS51 A0A0L7LU60 A0A089QAK4 A0A2A4K0W1 A0A3S2L6H3 B4X991 I6TMX7 A0A3S2NRK5 H9JG67 J7FDV3 Q56IC2 A0A2A4JSJ6 T1QDH5 T1QDG7 Q56IC3 A0A194RRT9 A0PGE0 D7RZZ8 B6CMF0 A0A194PGK9 D7RZZ6 D7RZZ7 D2Y4U4 D7RZZ9 E7D005 C7FF06 H9JG68 Q56IC4 Q86M89 A0A0L7LB31 A0A194PGV0 A0A194RV47 C9W8G0 J7FCZ8 A0A089Q9N5 A0A2A4J8W1 Q9N6C6 I6TEW9 O18450 O18444 E7D017 A0A2A4K197 A0A2A4K0D3 C9W8G8 E7D003 C9W8G7 B4X988 B4X987 A0A2A4K1Q8 A0A2A4K1C2 O18445 A0A1B0RHP3 A0A212F5Y7 B4X989 A0PGD7 A0A0K0Q479 A0A2A4K239 A0A2A4K228 A0A2A4J8H7 C9W8H8 A0A2A4J6X3 C9W8F9 O18443 A0A2A4K1R6 B4X990 E7D015 A0A2H1WAZ6 A9P3S5 A0A2A4J7U7 A0A2A4J6T8 A0PGD6 A0A142I706 E2D741 A0A0L7LAV4 A0A2H1V6V4 Q9NH08 A0A0X9HJY6 A0A1B1LTR9 A0A2H1VCC5 A0A212ES10 E7D007 Q9NH07 A0A2H1V6R8 Q25503 A0A1B1LTR7 I6TMX9 A0A2A4JTJ5 A0A1E1WDT0
A0A2A4K202 A0A2H1V6W6 A0A089QAL5 A0A0K8SR13 A0A2H1VX12 A0A0K8SS51 A0A0L7LU60 A0A089QAK4 A0A2A4K0W1 A0A3S2L6H3 B4X991 I6TMX7 A0A3S2NRK5 H9JG67 J7FDV3 Q56IC2 A0A2A4JSJ6 T1QDH5 T1QDG7 Q56IC3 A0A194RRT9 A0PGE0 D7RZZ8 B6CMF0 A0A194PGK9 D7RZZ6 D7RZZ7 D2Y4U4 D7RZZ9 E7D005 C7FF06 H9JG68 Q56IC4 Q86M89 A0A0L7LB31 A0A194PGV0 A0A194RV47 C9W8G0 J7FCZ8 A0A089Q9N5 A0A2A4J8W1 Q9N6C6 I6TEW9 O18450 O18444 E7D017 A0A2A4K197 A0A2A4K0D3 C9W8G8 E7D003 C9W8G7 B4X988 B4X987 A0A2A4K1Q8 A0A2A4K1C2 O18445 A0A1B0RHP3 A0A212F5Y7 B4X989 A0PGD7 A0A0K0Q479 A0A2A4K239 A0A2A4K228 A0A2A4J8H7 C9W8H8 A0A2A4J6X3 C9W8F9 O18443 A0A2A4K1R6 B4X990 E7D015 A0A2H1WAZ6 A9P3S5 A0A2A4J7U7 A0A2A4J6T8 A0PGD6 A0A142I706 E2D741 A0A0L7LAV4 A0A2H1V6V4 Q9NH08 A0A0X9HJY6 A0A1B1LTR9 A0A2H1VCC5 A0A212ES10 E7D007 Q9NH07 A0A2H1V6R8 Q25503 A0A1B1LTR7 I6TMX9 A0A2A4JTJ5 A0A1E1WDT0
Pubmed
EMBL
KX380994
ANS56510.1
FJ205401
ADM35105.1
FJ205400
ACR15969.2
+ More
NWSH01000291 PCG77701.1 ODYU01000971 SOQ36525.1 ODYU01004792 SOQ45041.1 PCG77700.1 SOQ36526.1 KM083794 AIR09775.1 GBRD01010002 JAG55822.1 SOQ45042.1 GBRD01009676 JAG56148.1 JTDY01000073 KOB79018.1 KM083784 AIR09765.1 PCG77699.1 RSAL01000119 RVE46739.1 EF531627 ABR88238.1 JQ904133 AFM77763.1 RVE46738.1 BABH01002097 JN252035 AFM28248.1 AY953055 AAX62028.1 NWSH01000651 PCG75015.1 JQ429441 KR024676 AFW03966.1 ALE15218.1 JQ429440 AFW03965.1 AY953054 AAX62027.1 KQ459765 KPJ20040.1 AY618895 AAV33658.1 HM209421 ADI32882.1 EU325550 ACB54940.1 KQ459604 KPI92526.1 HM209419 ADI32880.1 HM209420 ADI32881.1 GU323796 ADA83701.1 HM209422 ADI32883.1 HM990175 ADT80823.1 GQ354838 ACU00133.1 BABH01002098 AY953053 AAX62026.1 AY251276 AAO75039.1 JTDY01001894 KOB72615.1 KPI92527.1 KPJ20041.1 FJ205405 ACR15973.2 JN252046 AFM28259.1 KM083788 AIR09769.1 NWSH01002619 PCG67833.1 AF233731 AF233734 AAF71518.1 AAF71716.1 JQ904134 AFM77764.1 Y12287 CAA72966.1 Y12280 CAA72959.1 HM990187 ADT80835.1 PCG77698.1 PCG77697.1 FJ205413 ACR15981.1 HM990173 ADT80821.1 FJ205412 ACR15980.1 EF531624 ABR88235.1 EF531623 ABR88234.1 PCG77696.1 PCG77693.1 PCG77694.1 Y12281 CAA72960.1 KM360186 AKH49602.1 AGBW02010111 OWR49146.1 EF531625 ABR88236.1 AY618892 AAV33655.1 KP730444 AKR06192.1 PCG77702.1 PCG77692.1 PCG67834.1 FJ205424 ACR15991.1 PCG67835.1 FJ205404 ACR15972.2 Y12279 CAA72958.1 PCG77703.1 EF531626 ABR88237.1 HM990185 ADT80833.1 ODYU01007460 SOQ50255.1 EF104914 ABP96916.1 PCG67836.1 PCG67837.1 AY618891 AAV33654.1 KT907053 AMR44225.1 GQ891130 ADK27715.1 KOB72613.1 SOQ36529.1 AF233730 AAF71517.1 KP690081 ALO61083.1 SOQ36527.1 KX380993 ANS56509.1 ODYU01001778 SOQ38451.1 AGBW02012905 OWR44234.1 HM990177 ADT80825.1 AF233732 AAF71519.1 SOQ36528.1 L34168 AAA58743.1 KX380992 ANS56508.1 JQ904138 AFM77768.1 PCG75018.1 GDQN01005957 JAT85097.1
NWSH01000291 PCG77701.1 ODYU01000971 SOQ36525.1 ODYU01004792 SOQ45041.1 PCG77700.1 SOQ36526.1 KM083794 AIR09775.1 GBRD01010002 JAG55822.1 SOQ45042.1 GBRD01009676 JAG56148.1 JTDY01000073 KOB79018.1 KM083784 AIR09765.1 PCG77699.1 RSAL01000119 RVE46739.1 EF531627 ABR88238.1 JQ904133 AFM77763.1 RVE46738.1 BABH01002097 JN252035 AFM28248.1 AY953055 AAX62028.1 NWSH01000651 PCG75015.1 JQ429441 KR024676 AFW03966.1 ALE15218.1 JQ429440 AFW03965.1 AY953054 AAX62027.1 KQ459765 KPJ20040.1 AY618895 AAV33658.1 HM209421 ADI32882.1 EU325550 ACB54940.1 KQ459604 KPI92526.1 HM209419 ADI32880.1 HM209420 ADI32881.1 GU323796 ADA83701.1 HM209422 ADI32883.1 HM990175 ADT80823.1 GQ354838 ACU00133.1 BABH01002098 AY953053 AAX62026.1 AY251276 AAO75039.1 JTDY01001894 KOB72615.1 KPI92527.1 KPJ20041.1 FJ205405 ACR15973.2 JN252046 AFM28259.1 KM083788 AIR09769.1 NWSH01002619 PCG67833.1 AF233731 AF233734 AAF71518.1 AAF71716.1 JQ904134 AFM77764.1 Y12287 CAA72966.1 Y12280 CAA72959.1 HM990187 ADT80835.1 PCG77698.1 PCG77697.1 FJ205413 ACR15981.1 HM990173 ADT80821.1 FJ205412 ACR15980.1 EF531624 ABR88235.1 EF531623 ABR88234.1 PCG77696.1 PCG77693.1 PCG77694.1 Y12281 CAA72960.1 KM360186 AKH49602.1 AGBW02010111 OWR49146.1 EF531625 ABR88236.1 AY618892 AAV33655.1 KP730444 AKR06192.1 PCG77702.1 PCG77692.1 PCG67834.1 FJ205424 ACR15991.1 PCG67835.1 FJ205404 ACR15972.2 Y12279 CAA72958.1 PCG77703.1 EF531626 ABR88237.1 HM990185 ADT80833.1 ODYU01007460 SOQ50255.1 EF104914 ABP96916.1 PCG67836.1 PCG67837.1 AY618891 AAV33654.1 KT907053 AMR44225.1 GQ891130 ADK27715.1 KOB72613.1 SOQ36529.1 AF233730 AAF71517.1 KP690081 ALO61083.1 SOQ36527.1 KX380993 ANS56509.1 ODYU01001778 SOQ38451.1 AGBW02012905 OWR44234.1 HM990177 ADT80825.1 AF233732 AAF71519.1 SOQ36528.1 L34168 AAA58743.1 KX380992 ANS56508.1 JQ904138 AFM77768.1 PCG75018.1 GDQN01005957 JAT85097.1
Proteomes
Pfam
PF00089 Trypsin
Interpro
SUPFAM
SSF50494
SSF50494
CDD
ProteinModelPortal
A0A1B1LTS0
E0W9N4
C9W8F6
A0A2A4K0B4
A0A2H1V6P6
A0A2H1VW13
+ More
A0A2A4K202 A0A2H1V6W6 A0A089QAL5 A0A0K8SR13 A0A2H1VX12 A0A0K8SS51 A0A0L7LU60 A0A089QAK4 A0A2A4K0W1 A0A3S2L6H3 B4X991 I6TMX7 A0A3S2NRK5 H9JG67 J7FDV3 Q56IC2 A0A2A4JSJ6 T1QDH5 T1QDG7 Q56IC3 A0A194RRT9 A0PGE0 D7RZZ8 B6CMF0 A0A194PGK9 D7RZZ6 D7RZZ7 D2Y4U4 D7RZZ9 E7D005 C7FF06 H9JG68 Q56IC4 Q86M89 A0A0L7LB31 A0A194PGV0 A0A194RV47 C9W8G0 J7FCZ8 A0A089Q9N5 A0A2A4J8W1 Q9N6C6 I6TEW9 O18450 O18444 E7D017 A0A2A4K197 A0A2A4K0D3 C9W8G8 E7D003 C9W8G7 B4X988 B4X987 A0A2A4K1Q8 A0A2A4K1C2 O18445 A0A1B0RHP3 A0A212F5Y7 B4X989 A0PGD7 A0A0K0Q479 A0A2A4K239 A0A2A4K228 A0A2A4J8H7 C9W8H8 A0A2A4J6X3 C9W8F9 O18443 A0A2A4K1R6 B4X990 E7D015 A0A2H1WAZ6 A9P3S5 A0A2A4J7U7 A0A2A4J6T8 A0PGD6 A0A142I706 E2D741 A0A0L7LAV4 A0A2H1V6V4 Q9NH08 A0A0X9HJY6 A0A1B1LTR9 A0A2H1VCC5 A0A212ES10 E7D007 Q9NH07 A0A2H1V6R8 Q25503 A0A1B1LTR7 I6TMX9 A0A2A4JTJ5 A0A1E1WDT0
A0A2A4K202 A0A2H1V6W6 A0A089QAL5 A0A0K8SR13 A0A2H1VX12 A0A0K8SS51 A0A0L7LU60 A0A089QAK4 A0A2A4K0W1 A0A3S2L6H3 B4X991 I6TMX7 A0A3S2NRK5 H9JG67 J7FDV3 Q56IC2 A0A2A4JSJ6 T1QDH5 T1QDG7 Q56IC3 A0A194RRT9 A0PGE0 D7RZZ8 B6CMF0 A0A194PGK9 D7RZZ6 D7RZZ7 D2Y4U4 D7RZZ9 E7D005 C7FF06 H9JG68 Q56IC4 Q86M89 A0A0L7LB31 A0A194PGV0 A0A194RV47 C9W8G0 J7FCZ8 A0A089Q9N5 A0A2A4J8W1 Q9N6C6 I6TEW9 O18450 O18444 E7D017 A0A2A4K197 A0A2A4K0D3 C9W8G8 E7D003 C9W8G7 B4X988 B4X987 A0A2A4K1Q8 A0A2A4K1C2 O18445 A0A1B0RHP3 A0A212F5Y7 B4X989 A0PGD7 A0A0K0Q479 A0A2A4K239 A0A2A4K228 A0A2A4J8H7 C9W8H8 A0A2A4J6X3 C9W8F9 O18443 A0A2A4K1R6 B4X990 E7D015 A0A2H1WAZ6 A9P3S5 A0A2A4J7U7 A0A2A4J6T8 A0PGD6 A0A142I706 E2D741 A0A0L7LAV4 A0A2H1V6V4 Q9NH08 A0A0X9HJY6 A0A1B1LTR9 A0A2H1VCC5 A0A212ES10 E7D007 Q9NH07 A0A2H1V6R8 Q25503 A0A1B1LTR7 I6TMX9 A0A2A4JTJ5 A0A1E1WDT0
PDB
2HLC
E-value=1.32729e-29,
Score=322
Ontologies
GO
Topology
SignalP
Position: 1 - 15,
Likelihood: 0.939199
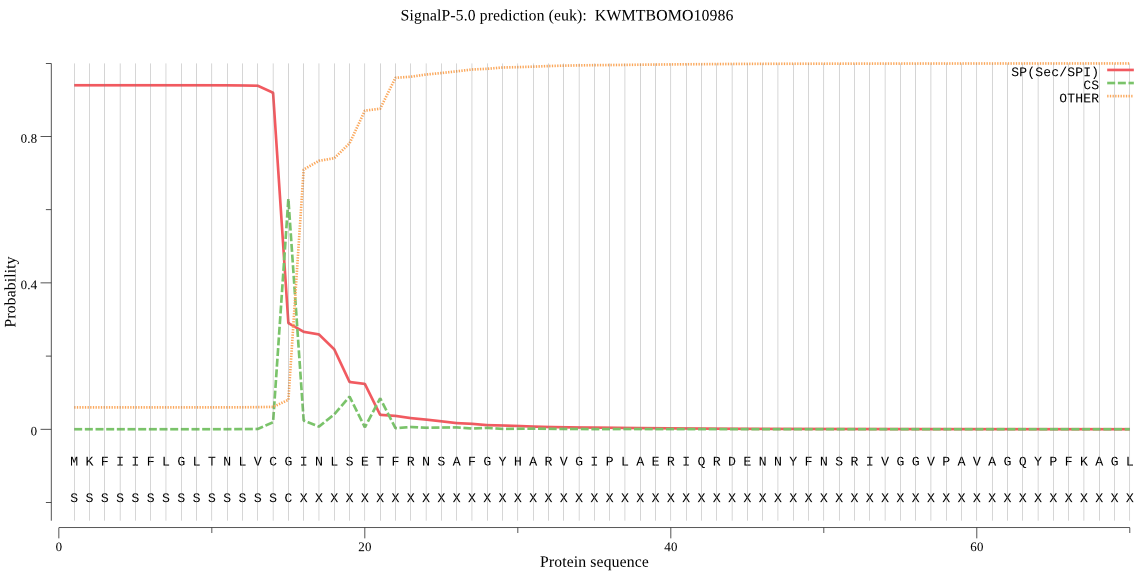
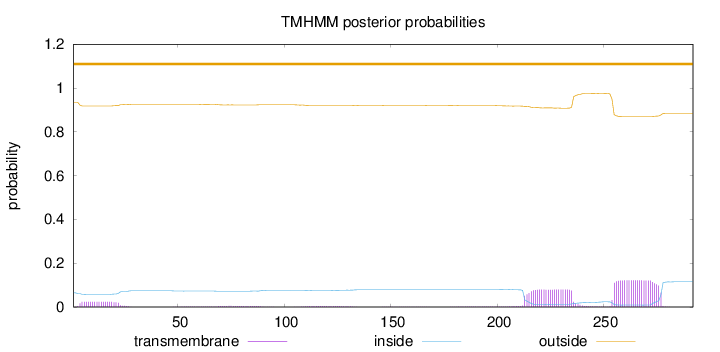
Length:
292
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
5.29252
Exp number, first 60 AAs:
0.48397
Total prob of N-in:
0.06526
outside
1 - 292
Population Genetic Test Statistics
Pi
165.323936
Theta
162.230749
Tajima's D
0.183317
CLR
24.144975
CSRT
0.418329083545823
Interpretation
Uncertain
