Gene
KWMTBOMO10978
Annotation
reverse_transcriptase_[Danaus_plexippus]
Location in the cell
Mitochondrial Reliability : 1.386 Nuclear Reliability : 1.055 PlasmaMembrane Reliability : 1.07
Sequence
CDS
ATGCTCACGAATCAATATACTCGTAGAGAATATCATAACAAAAGAATCAGATATAAATTCGTAATTACGCAACGCTATCCTGATCACACAGGAGCCAGTCACCGTCTACGCCCTAGACACGTCCTTACGGATCCATCAGATCCAATAACCTTTGCATTAGACGCCTTCAGCTCTAACACTAGAAGTGGCCTGGACGACTGCACCACCAGGGATGAGGTGACGGCGGCCGTCGCTGCCCAGGGCAAATGCGCACTGGAAAATGTTACAGTAGGCGAGCCGCGCCGCGGTTATACCGGGGCCAGCTCGGTATGGGTGCGTTGCCCGGTGGAAGCGGCCGCCTCCCTGGCTGCTCCTCCTCCCGGGAGACCTTCGGGAAATCCGGGCATGCTGCGCGTGGGCTGGATAGCGGCCCACGTGCACTTGCTTGAACCACGCGCCTGGCGATGCCTCCGGTGCTTTAGCACCGGACACGGCCTCGCCAGGTGCACCAGCTCGGTTGACCGCAGTGGTCTGTGTTTCCGCTGCGGCCAGCCGGGACATAAAGCGGCGTCCTGCACGGCTGCCCCGCATTGCGCTCTGTGTGCTGCGGCCGGGCGCAGGGCAAGCCACCGAACGGGAGGCAAGGCGTGCTTTTCGTCCGGTAATAATACCGAACGCAACCGGCGCCGCAAGTCAAAGCGTCGGCAAAACAGAAGGTTGGCCAGAGGAGCACTGGCCAACGCTGTAATCCCCGCTGCGGTGCCGGTGCCGGAGGGCGGGGGCAGTGGTGAAGGGGAGGGGGCGATGGATGACATGCTGTCTCAGAGCATGGCGGAGTGGTCCATAGATGTGGCTGTGGTCGCCGAGCCGTACTTCGTTCCCCCGGCAAATGATTGCTGGTTTGGGGACGATGGCGGCCTGGCGGCCATAGTCATAAGAAGAGACGCGGCGATCCCCCCCCTGGCGATGGTGACCAGGGGCCAAGGTTTCGTCGCGGTGCAGTTGGAAGGGACCGTCGTGATCGGGGCGTACTTCTCTCCAAACAGGCCTACTGTCGAGTTCGGGCATTTCCTGGGCGGGATCGGGGCGATTGCTCGCCGCCTCGCTCCCCGTCCAGTGATTCTCGCGGGGGACCTCAATGCGAAGTCTGTCGCATGGGGTTCCCCCCGCTCGGACGCTCGTGGTAGGCTGCTGGAGAGGTGGGCGAACGTGGCCGGCCTCTGTTTGTTGAATAGAGGTTCGGTTGCGACGTGCGTGCGGTGGCAGGGCGAGTCTATTGTGGACGTTACGTTCGCAAGCCCGGCCATCGCGCGCCGTATCCGCGACTGGAGGGTCTTGGAGGGGGCGGAGACACTGTCAGATCACCGATATATTCGGTTTGATCTCTCCGCCCGCTCCGCAGTCGCGGACGTCCACAGGGACCACCCCCATGGTGCGTCCCGGTCATTCCCCAAGTGGGCACTGAAGCGCCTGAATAAGGAGCTCCTGATGGAAGCCTCTATGGTGGCGGCGTGGGCGCCCATGCGTCCACACCCGGTCGAGGTCGAGGGCGAGGCAGGGTGGTTCCGGGACACGATGTGTCGCATTTGTGATGCGTCGATGCCCCGGATTGGCCCTCGACTTCCTAATCGCCAAGCGTACTGGTGGTCGCCCGAAATTGCGCAACTCCGCGTGGAGTGCGTTCGGGCGAGGCGCGAGTGCGCCAGGCATCGCCGCCGCCGCCTGCCGCGACGCAATGATCCGGTTGCATTCGCGGCGGAAGAGGTCCGGCTGCACGGCGCATTTCGCGTCGCGCAGAGGGCATTGCAGCTGGCCATCAGAAGAGCCAAGAACCAACACATGGAGGGTCTCTTGGAGACGCTGGATGAGGATCCGTGGGGGCGGCCCTATCATATGGTGCGCAATAAAATGCGCCCATGGGCCCCCCCGATCACGGAGCGTCTCCAGCCTCAGCAGCTGCGGGACATCGTCTCGGCGCTGTTCCCGCAGGAGCGGGATGGATTTATCGCTCCCGCTATGGACACGCCGCCGGAATACGACGGTGAAGTCCCTGCTGAGGTGCCCCATATAACGGGGGCGGAGCTCCGTGTGGCCGTACGCAAAATGTGCGCGAAGGACACTGCACCCGGCCCGGACGGCGTCCATGGTCGGGTTTGGGGCTTGGCTCTTGGTGCCCTGGGGGACCGACTCTTGAGGCTTTACAACTCCTGCCTCGAGTCGGGACGGTTTCCTTCATCGTGGAGGACGGGCAGACTCGTGCTGTTGAGAAAGGAGGGGCGCCCGGCGGATACCGCCGCCGGGTACCGTCCCATCGTGTTGCTGGATGAGGTGGGCAAGCTGCTGGAACGTATTCTGGCAGCCCGCATCATCCAGCATCTGGCCGGGGTGGGACCCGATCTGTCGGCGGAACAGTATGGCTTCCGAGAGGGCCGCTCAACCGTAGACGCGATCCTTCGCGTGCGGGCCCTCTCGGAGGAGGCCGTCTCCAGGGGCGGGGTGGCCCTGGCGGTATCACTCGATATCGCCAATGCGTTCAACACCCTGCCCTGGTCCGTGATAGGGGGGCGCTGGAAAGACACGGAGTGCCCCCCTACCTTCGCCGGCTGGCCCCGGAGAGTGCCACCTATCGATGCTCAAATCGTGGTTGGAGGCGTCCGTATTGGGGTCGGGGTGCAGTTGAAGTACCTCGGCCTCATTCTAGACAGTCGTTGGACCTTCCGTGCTCACTTTCAGAATCTGGTCCCTCGTTTGTTGGGGGTGGCCGGCGCGTTAAGCCGGCTTCTGCCCAACGTCGGGGGGCCTGACCAGGTGACGCGCCGTCTCTATACAGGGGTGGTGCGGTCAATGGCCCTATACGGGGCGCCCGTGTGGGGCCAGTCCCTGGCCGTGGGGGTAGCAAAGTTGCTGCAACGGCCGCAACGCACCATCGCGGTCAGGGTCATCCGCGGCTATCGCACCATCTCCTTCGAGGCGGCGTGTGTACTGGCTGGGACGCCGCCTTGGGTTCTGGAAGCGGAGGCGCTCGCCGCTGACTATCAGTGGCGGGCTGACTTTCGTGCCCGGGGCGTGGCGCGCCCCAGCCCCAGTGTGGTCAGAGCGCGGAGGGCCCAATCTCGGCGGTCCGTGCTGGAATCATGGTCCAGGCGGCTGGCCGATCCTTCGGCTGGTCGTAGGACCGTCGAGGCGATTCGCCCAGTCCTTGTGGACTGGGCGAATCGTGACAGAGGACGCCTCACTTTCCGGCTCACGCAGGTGCTCACTGGGCATGGTTGCTTCGGTGAGTTCCTGCACCGGATCAGAGCCGAGCCGACGGCAGAGTGCCACCATTGTGGTTGCGACTTGGACACGGCAGAGCACACGCTCGTTGCCTGCCCCGCATGGGAGGGGTGGCGCCGTGTCCTCGTCGCAAAAATAGGAAACGACTTGTCGTTGCCGAGTGTTGTGGCATCGATGCTCGGCGACGACGAGTCGTGGAAGGCGATGCTCGACTTCTGCGAGTGCACCATCTCGCAGAAGGAGGCGGCGGGGCGCGTGAGAGACGCGCAAGCCCGCCGCCGTCGAGCGGGGGCCAGGGAGGCGGATATCGCCCAAGCCCTGGCCCTCTAA
Protein
MLTNQYTRREYHNKRIRYKFVITQRYPDHTGASHRLRPRHVLTDPSDPITFALDAFSSNTRSGLDDCTTRDEVTAAVAAQGKCALENVTVGEPRRGYTGASSVWVRCPVEAAASLAAPPPGRPSGNPGMLRVGWIAAHVHLLEPRAWRCLRCFSTGHGLARCTSSVDRSGLCFRCGQPGHKAASCTAAPHCALCAAAGRRASHRTGGKACFSSGNNTERNRRRKSKRRQNRRLARGALANAVIPAAVPVPEGGGSGEGEGAMDDMLSQSMAEWSIDVAVVAEPYFVPPANDCWFGDDGGLAAIVIRRDAAIPPLAMVTRGQGFVAVQLEGTVVIGAYFSPNRPTVEFGHFLGGIGAIARRLAPRPVILAGDLNAKSVAWGSPRSDARGRLLERWANVAGLCLLNRGSVATCVRWQGESIVDVTFASPAIARRIRDWRVLEGAETLSDHRYIRFDLSARSAVADVHRDHPHGASRSFPKWALKRLNKELLMEASMVAAWAPMRPHPVEVEGEAGWFRDTMCRICDASMPRIGPRLPNRQAYWWSPEIAQLRVECVRARRECARHRRRRLPRRNDPVAFAAEEVRLHGAFRVAQRALQLAIRRAKNQHMEGLLETLDEDPWGRPYHMVRNKMRPWAPPITERLQPQQLRDIVSALFPQERDGFIAPAMDTPPEYDGEVPAEVPHITGAELRVAVRKMCAKDTAPGPDGVHGRVWGLALGALGDRLLRLYNSCLESGRFPSSWRTGRLVLLRKEGRPADTAAGYRPIVLLDEVGKLLERILAARIIQHLAGVGPDLSAEQYGFREGRSTVDAILRVRALSEEAVSRGGVALAVSLDIANAFNTLPWSVIGGRWKDTECPPTFAGWPRRVPPIDAQIVVGGVRIGVGVQLKYLGLILDSRWTFRAHFQNLVPRLLGVAGALSRLLPNVGGPDQVTRRLYTGVVRSMALYGAPVWGQSLAVGVAKLLQRPQRTIAVRVIRGYRTISFEAACVLAGTPPWVLEAEALAADYQWRADFRARGVARPSPSVVRARRAQSRRSVLESWSRRLADPSAGRRTVEAIRPVLVDWANRDRGRLTFRLTQVLTGHGCFGEFLHRIRAEPTAECHHCGCDLDTAEHTLVACPAWEGWRRVLVAKIGNDLSLPSVVASMLGDDESWKAMLDFCECTISQKEAAGRVRDAQARRRRAGAREADIAQALAL
Summary
Uniprot
ProteinModelPortal
PDB
1WDU
E-value=5.39705e-11,
Score=167
Ontologies
GO
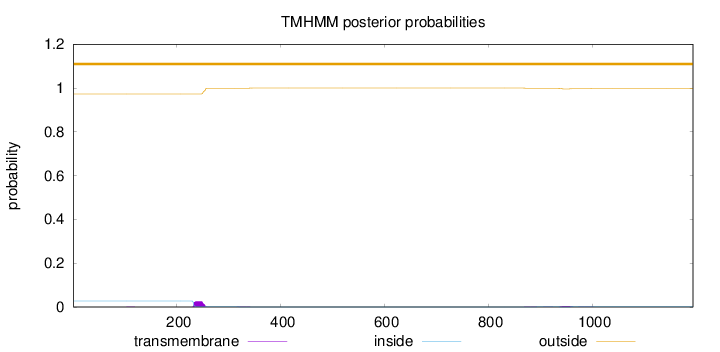
Topology
Length:
1194
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.677399999999997
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02740
outside
1 - 1194
Population Genetic Test Statistics
Pi
132.672335
Theta
24.67782
Tajima's D
0.67003
CLR
0.213053
CSRT
0.574121293935303
Interpretation
Uncertain
