Gene
KWMTBOMO10972
Annotation
UDP-glycosyltransferase_UGT44A1_[Bombyx_mori]
Full name
UDP-glucuronosyltransferase
Location in the cell
PlasmaMembrane Reliability : 1.48
Sequence
CDS
ATGTCAGATGCTCCGGCTTCGTACAGAGACATTTTATTAAGTGACAGACATGTGTATAAAGGTTTATCTTTTGAGTCAGTGATAGCAAGTGAAGTGTCGCGAGTGCCCTTTGAAACACTAGTGGCGACAAAAGCCGGCAACGATGATTGCAAAACTCTCATGAATAATAATCAAGTACTGCATTTGATTAGGACGCGGCCTCAATATGACGTCGTACTGGTCGAATCATTTAACAGTGACTGCGGAATAGCATTGGCGGCAAATTTGAGTGCGCCGTACATAGCATTGAACCCGAAGCCTTTACAGCCTTGGCACTACAATCGATTAGGTATCAATTTCAATGCAGCTTATGTAACCCAAACCGGCTTATCGTACGGAAAAAATCCTTGGTTCCTAGACCGAGTAAGAGGCTACATATTGTATCATATTACAAACTGGGTGTACTATGTTGGTTCACAAATCACAGATCACGTGTACTTATATAAATATTTAGGAGACAACCTTCCGTCTCTCGAAACGCTTGCGTCAAATGCTAGTTTAGTGTTTGTAAACACACATCAATCCGTTTTTGGGGGTATCTCCAGGCCTGATAACGTAATAGATATTGGCGGCATACACGTGAGACCTCCAAAAATCATTCCAACAGAAATCGAGAGATTTATAAACGAGGCTCAGCATGGTGTCGTCTATGTGAACTTGGGTTCAACGGTAAAAGATTCCACCCTGCCTGCCGAAAAGCTGGCTGAACTCCTCCTCACCTTCAGAAAACTACCCCACAGGGTACTTTGGAAATGGGACGGAGCAGCCATACAGAACCTGCCAAGAAATGTTATGACAATGAAGTGGCTACCCCAATATGACATTTTAAAGCACAAAAATGTAAAAGCACTCATCACTCATGCCGGTATACTGAGCACCATTGAAGCTATCGATGCTGGTATCCCTGTGGTGGCTATTCCCTTATTTGGAGATCAATATGGCAATGCAGCAGCAATGCAAGACGCCGGCATGGCGACCATAGTGCATTACCAGGATTTAAATAAAGAACATCTACTGGGTGCAGTCAATGAAGTTTTGGATGCGAAGAGGCAACAACAAGCAAAGCTGACGTCCAGGTTGTGGCACGACCGTTCTTTGTCGCCTTTGGAAAACGCCATCTATTGGACGGAATACGTAGCTCGATACCAAGGAGCTCCCAATCTTCAGCCTTTGTCGTCACAAGCACCACTATATCAACAATTGCAGCTAGACGTGTTATTGTTTGTGGCAATCGTTGTTTATATCCTGTTCTACGCTTTGTACAAAATCCTCAGAACCCTTTGCTGTTGTTGCTGTCGAGCTGATTCCGGAAACGGCGATGACGTAGGGGACACAAGGAAAAGAAAAAGAGTAAAATTCGAATGA
Protein
MSDAPASYRDILLSDRHVYKGLSFESVIASEVSRVPFETLVATKAGNDDCKTLMNNNQVLHLIRTRPQYDVVLVESFNSDCGIALAANLSAPYIALNPKPLQPWHYNRLGINFNAAYVTQTGLSYGKNPWFLDRVRGYILYHITNWVYYVGSQITDHVYLYKYLGDNLPSLETLASNASLVFVNTHQSVFGGISRPDNVIDIGGIHVRPPKIIPTEIERFINEAQHGVVYVNLGSTVKDSTLPAEKLAELLLTFRKLPHRVLWKWDGAAIQNLPRNVMTMKWLPQYDILKHKNVKALITHAGILSTIEAIDAGIPVVAIPLFGDQYGNAAAMQDAGMATIVHYQDLNKEHLLGAVNEVLDAKRQQQAKLTSRLWHDRSLSPLENAIYWTEYVARYQGAPNLQPLSSQAPLYQQLQLDVLLFVAIVVYILFYALYKILRTLCCCCCRADSGNGDDVGDTRKRKRVKFE
Summary
Catalytic Activity
glucuronate acceptor + UDP-alpha-D-glucuronate = acceptor beta-D-glucuronoside + H(+) + UDP
Similarity
Belongs to the UDP-glycosyltransferase family.
Feature
chain UDP-glucuronosyltransferase
Uniprot
G9LPV9
G9LPR5
A0A2A4JHL4
A0A3S2PBM0
A0A194PN21
A0A191T1N3
+ More
A0A286MXP5 A0A2H1W091 A0A194RRT3 A0A0L7LRH5 A0A2H4WB74 A0A1L8D6F8 A0A212EWF8 S4PCR1 A0A3S2LHF3 A0A2J7R4V4 A0A2H4WB56 G9LPV6 D2A2V5 G9LPR2 A0A2J7R4W2 A0A2J7R4W8 A0A2A4JZV9 A0A194PHJ2 A0A1L8D6F4 A0A212EWE0 A0A0L7RH14 I4DM95 A0A2J7R4S8 A0A1Y1LIT5 A0A2H4WB57 A0A0A1WW96 A0A191T2H3 A0A191T1P6 G9LPV5 A0A067RAD1 A0A024AHN3 V5H2B2 A0A2H4WB66 A0A2P8ZG71 A0A087ZYR9 A0A2P8ZG63 A0A286MXP4 A0A0K8UGE5 A0A212EWH2 A0A0K8UM10 G9LPV7 A0A2P8ZG78 A0A182NE44 A0A194RRD0 A0A182PJ64 A0A1W4WYE4 A0A182W9Q0 A0A1I8NWG2 A0A182UEM4 A0A182V6Z3 A0A1S4GXF8 A0A182WT93 J9K7F3 A0A1Y1LL43 G9LPR3 A0A1W4WMF1 A0A182HV86 A0A2J7R4W5 A0A182YGV2 A0A1B6KV12 B3NL92 A0A1L8D6L6 B4JDJ1 A0A182RBM8 A0A1I8NWF4 W8AR85 A0A2J7R4T0 A0A067QGQ4 A0A2P8ZG70 A0A194RRL0 A0A1I8N2C0 T1PFI1 Q9VJ46 D6W4K8 A0A0J9R3P5 B4Q8Y7 A0A1W4VM86 A0A1W4V9G2 A0A1W4VMW4 B4G9A2 A0A139WIW7 B4G9A5 A0A084VP43 A0A182QSB1 A0A1W4V9E9 E2B0N7 A0A0C9RNU1 B4I5L1 A0A026WYL1 Q29MG5 A0A1W4WMC9 A0A034V788 A0A291PQF1 B4Q8Y8
A0A286MXP5 A0A2H1W091 A0A194RRT3 A0A0L7LRH5 A0A2H4WB74 A0A1L8D6F8 A0A212EWF8 S4PCR1 A0A3S2LHF3 A0A2J7R4V4 A0A2H4WB56 G9LPV6 D2A2V5 G9LPR2 A0A2J7R4W2 A0A2J7R4W8 A0A2A4JZV9 A0A194PHJ2 A0A1L8D6F4 A0A212EWE0 A0A0L7RH14 I4DM95 A0A2J7R4S8 A0A1Y1LIT5 A0A2H4WB57 A0A0A1WW96 A0A191T2H3 A0A191T1P6 G9LPV5 A0A067RAD1 A0A024AHN3 V5H2B2 A0A2H4WB66 A0A2P8ZG71 A0A087ZYR9 A0A2P8ZG63 A0A286MXP4 A0A0K8UGE5 A0A212EWH2 A0A0K8UM10 G9LPV7 A0A2P8ZG78 A0A182NE44 A0A194RRD0 A0A182PJ64 A0A1W4WYE4 A0A182W9Q0 A0A1I8NWG2 A0A182UEM4 A0A182V6Z3 A0A1S4GXF8 A0A182WT93 J9K7F3 A0A1Y1LL43 G9LPR3 A0A1W4WMF1 A0A182HV86 A0A2J7R4W5 A0A182YGV2 A0A1B6KV12 B3NL92 A0A1L8D6L6 B4JDJ1 A0A182RBM8 A0A1I8NWF4 W8AR85 A0A2J7R4T0 A0A067QGQ4 A0A2P8ZG70 A0A194RRL0 A0A1I8N2C0 T1PFI1 Q9VJ46 D6W4K8 A0A0J9R3P5 B4Q8Y7 A0A1W4VM86 A0A1W4V9G2 A0A1W4VMW4 B4G9A2 A0A139WIW7 B4G9A5 A0A084VP43 A0A182QSB1 A0A1W4V9E9 E2B0N7 A0A0C9RNU1 B4I5L1 A0A026WYL1 Q29MG5 A0A1W4WMC9 A0A034V788 A0A291PQF1 B4Q8Y8
EC Number
2.4.1.17
Pubmed
22155036
26354079
19954531
25299618
26227816
29027758
+ More
22118469 23622113 18362917 19820115 22651552 28004739 25830018 24845553 24698447 29403074 12364791 25244985 17994087 24495485 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 24438588 20798317 24508170 15632085 25348373
22118469 23622113 18362917 19820115 22651552 28004739 25830018 24845553 24698447 29403074 12364791 25244985 17994087 24495485 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 24438588 20798317 24508170 15632085 25348373
EMBL
JQ070266
AEW43182.1
JQ070222
AEW43138.1
NWSH01001552
PCG70882.1
+ More
RSAL01000119 RVE46733.1 KQ459604 KPI92520.1 KU680308 ANI22014.1 MF684363 ASX93997.1 ODYU01005418 SOQ46242.1 KQ459765 KPJ20035.1 JTDY01000310 KOB77761.1 KY202936 AUC64280.1 GEYN01000098 JAV02031.1 AGBW02012000 OWR45816.1 GAIX01004036 JAA88524.1 RVE46732.1 NEVH01007402 PNF35866.1 KY202929 AUC64273.1 JQ070263 AEW43179.1 KQ971338 EFA02977.2 JQ070219 AEW43135.1 PNF35872.1 PNF35870.1 NWSH01000347 PCG77184.1 KPI92518.1 GEYN01000096 JAV02033.1 OWR45818.1 KQ414596 KOC70104.1 AK402413 BAM19035.1 PNF35836.1 GEZM01054555 JAV73559.1 KY202930 AUC64274.1 GBXI01011391 JAD02901.1 KU680305 ANI22011.1 KU680306 ANI22012.1 JQ070262 AEW43178.1 KK852590 KDR20744.1 KF777118 AHY99689.1 GALX01001513 JAB66953.1 KY202934 AUC64278.1 PYGN01000068 PSN55491.1 PSN55493.1 MF684362 ASX93996.1 GDHF01026592 JAI25722.1 OWR45817.1 GDHF01024708 JAI27606.1 JQ070264 AEW43180.1 PSN55501.1 KPJ20032.1 AAAB01008964 ABLF02020524 ABLF02020525 GEZM01054556 JAV73558.1 JQ070220 AEW43136.1 APCN01002501 PNF35871.1 GEBQ01024697 JAT15280.1 CH954179 EDV54742.1 GEYN01000094 JAV02035.1 CH916368 EDW03361.1 GAMC01019493 GAMC01019492 GAMC01019491 JAB87063.1 PNF35837.1 KK853490 KDR07235.1 PSN55494.1 KPJ20034.1 KA647552 AFP62181.1 AE014134 AY051629 AAF53709.1 AAK93053.1 BT124908 ADH29876.1 CM002910 KMY90882.1 CM000361 EDX05419.1 KMY90880.1 CH479180 EDW28932.1 KYB27856.1 EDW28935.1 ATLV01014974 KE524999 KFB39737.1 AXCN02000871 GL444666 EFN60709.1 GBYB01008756 JAG78523.1 CH480822 EDW55667.1 KK107063 EZA61120.1 CH379060 EAL33728.1 GAKP01020965 JAC37987.1 KY860727 ATL15306.1 EDX05420.1 KMY90883.1
RSAL01000119 RVE46733.1 KQ459604 KPI92520.1 KU680308 ANI22014.1 MF684363 ASX93997.1 ODYU01005418 SOQ46242.1 KQ459765 KPJ20035.1 JTDY01000310 KOB77761.1 KY202936 AUC64280.1 GEYN01000098 JAV02031.1 AGBW02012000 OWR45816.1 GAIX01004036 JAA88524.1 RVE46732.1 NEVH01007402 PNF35866.1 KY202929 AUC64273.1 JQ070263 AEW43179.1 KQ971338 EFA02977.2 JQ070219 AEW43135.1 PNF35872.1 PNF35870.1 NWSH01000347 PCG77184.1 KPI92518.1 GEYN01000096 JAV02033.1 OWR45818.1 KQ414596 KOC70104.1 AK402413 BAM19035.1 PNF35836.1 GEZM01054555 JAV73559.1 KY202930 AUC64274.1 GBXI01011391 JAD02901.1 KU680305 ANI22011.1 KU680306 ANI22012.1 JQ070262 AEW43178.1 KK852590 KDR20744.1 KF777118 AHY99689.1 GALX01001513 JAB66953.1 KY202934 AUC64278.1 PYGN01000068 PSN55491.1 PSN55493.1 MF684362 ASX93996.1 GDHF01026592 JAI25722.1 OWR45817.1 GDHF01024708 JAI27606.1 JQ070264 AEW43180.1 PSN55501.1 KPJ20032.1 AAAB01008964 ABLF02020524 ABLF02020525 GEZM01054556 JAV73558.1 JQ070220 AEW43136.1 APCN01002501 PNF35871.1 GEBQ01024697 JAT15280.1 CH954179 EDV54742.1 GEYN01000094 JAV02035.1 CH916368 EDW03361.1 GAMC01019493 GAMC01019492 GAMC01019491 JAB87063.1 PNF35837.1 KK853490 KDR07235.1 PSN55494.1 KPJ20034.1 KA647552 AFP62181.1 AE014134 AY051629 AAF53709.1 AAK93053.1 BT124908 ADH29876.1 CM002910 KMY90882.1 CM000361 EDX05419.1 KMY90880.1 CH479180 EDW28932.1 KYB27856.1 EDW28935.1 ATLV01014974 KE524999 KFB39737.1 AXCN02000871 GL444666 EFN60709.1 GBYB01008756 JAG78523.1 CH480822 EDW55667.1 KK107063 EZA61120.1 CH379060 EAL33728.1 GAKP01020965 JAC37987.1 KY860727 ATL15306.1 EDX05420.1 KMY90883.1
Proteomes
UP000218220
UP000283053
UP000053268
UP000053240
UP000037510
UP000007151
+ More
UP000235965 UP000007266 UP000053825 UP000027135 UP000245037 UP000005203 UP000075884 UP000075885 UP000192223 UP000075920 UP000095300 UP000075902 UP000075903 UP000076407 UP000007819 UP000075840 UP000076408 UP000008711 UP000001070 UP000075900 UP000095301 UP000000803 UP000000304 UP000192221 UP000008744 UP000030765 UP000075886 UP000000311 UP000001292 UP000053097 UP000001819
UP000235965 UP000007266 UP000053825 UP000027135 UP000245037 UP000005203 UP000075884 UP000075885 UP000192223 UP000075920 UP000095300 UP000075902 UP000075903 UP000076407 UP000007819 UP000075840 UP000076408 UP000008711 UP000001070 UP000075900 UP000095301 UP000000803 UP000000304 UP000192221 UP000008744 UP000030765 UP000075886 UP000000311 UP000001292 UP000053097 UP000001819
Pfam
PF00201 UDPGT
ProteinModelPortal
G9LPV9
G9LPR5
A0A2A4JHL4
A0A3S2PBM0
A0A194PN21
A0A191T1N3
+ More
A0A286MXP5 A0A2H1W091 A0A194RRT3 A0A0L7LRH5 A0A2H4WB74 A0A1L8D6F8 A0A212EWF8 S4PCR1 A0A3S2LHF3 A0A2J7R4V4 A0A2H4WB56 G9LPV6 D2A2V5 G9LPR2 A0A2J7R4W2 A0A2J7R4W8 A0A2A4JZV9 A0A194PHJ2 A0A1L8D6F4 A0A212EWE0 A0A0L7RH14 I4DM95 A0A2J7R4S8 A0A1Y1LIT5 A0A2H4WB57 A0A0A1WW96 A0A191T2H3 A0A191T1P6 G9LPV5 A0A067RAD1 A0A024AHN3 V5H2B2 A0A2H4WB66 A0A2P8ZG71 A0A087ZYR9 A0A2P8ZG63 A0A286MXP4 A0A0K8UGE5 A0A212EWH2 A0A0K8UM10 G9LPV7 A0A2P8ZG78 A0A182NE44 A0A194RRD0 A0A182PJ64 A0A1W4WYE4 A0A182W9Q0 A0A1I8NWG2 A0A182UEM4 A0A182V6Z3 A0A1S4GXF8 A0A182WT93 J9K7F3 A0A1Y1LL43 G9LPR3 A0A1W4WMF1 A0A182HV86 A0A2J7R4W5 A0A182YGV2 A0A1B6KV12 B3NL92 A0A1L8D6L6 B4JDJ1 A0A182RBM8 A0A1I8NWF4 W8AR85 A0A2J7R4T0 A0A067QGQ4 A0A2P8ZG70 A0A194RRL0 A0A1I8N2C0 T1PFI1 Q9VJ46 D6W4K8 A0A0J9R3P5 B4Q8Y7 A0A1W4VM86 A0A1W4V9G2 A0A1W4VMW4 B4G9A2 A0A139WIW7 B4G9A5 A0A084VP43 A0A182QSB1 A0A1W4V9E9 E2B0N7 A0A0C9RNU1 B4I5L1 A0A026WYL1 Q29MG5 A0A1W4WMC9 A0A034V788 A0A291PQF1 B4Q8Y8
A0A286MXP5 A0A2H1W091 A0A194RRT3 A0A0L7LRH5 A0A2H4WB74 A0A1L8D6F8 A0A212EWF8 S4PCR1 A0A3S2LHF3 A0A2J7R4V4 A0A2H4WB56 G9LPV6 D2A2V5 G9LPR2 A0A2J7R4W2 A0A2J7R4W8 A0A2A4JZV9 A0A194PHJ2 A0A1L8D6F4 A0A212EWE0 A0A0L7RH14 I4DM95 A0A2J7R4S8 A0A1Y1LIT5 A0A2H4WB57 A0A0A1WW96 A0A191T2H3 A0A191T1P6 G9LPV5 A0A067RAD1 A0A024AHN3 V5H2B2 A0A2H4WB66 A0A2P8ZG71 A0A087ZYR9 A0A2P8ZG63 A0A286MXP4 A0A0K8UGE5 A0A212EWH2 A0A0K8UM10 G9LPV7 A0A2P8ZG78 A0A182NE44 A0A194RRD0 A0A182PJ64 A0A1W4WYE4 A0A182W9Q0 A0A1I8NWG2 A0A182UEM4 A0A182V6Z3 A0A1S4GXF8 A0A182WT93 J9K7F3 A0A1Y1LL43 G9LPR3 A0A1W4WMF1 A0A182HV86 A0A2J7R4W5 A0A182YGV2 A0A1B6KV12 B3NL92 A0A1L8D6L6 B4JDJ1 A0A182RBM8 A0A1I8NWF4 W8AR85 A0A2J7R4T0 A0A067QGQ4 A0A2P8ZG70 A0A194RRL0 A0A1I8N2C0 T1PFI1 Q9VJ46 D6W4K8 A0A0J9R3P5 B4Q8Y7 A0A1W4VM86 A0A1W4V9G2 A0A1W4VMW4 B4G9A2 A0A139WIW7 B4G9A5 A0A084VP43 A0A182QSB1 A0A1W4V9E9 E2B0N7 A0A0C9RNU1 B4I5L1 A0A026WYL1 Q29MG5 A0A1W4WMC9 A0A034V788 A0A291PQF1 B4Q8Y8
PDB
2O6L
E-value=3.18996e-23,
Score=269
Ontologies
PATHWAY
00040
Pentose and glucuronate interconversions - Bombyx mori (domestic silkworm)
00053 Ascorbate and aldarate metabolism - Bombyx mori (domestic silkworm)
00830 Retinol metabolism - Bombyx mori (domestic silkworm)
00860 Porphyrin and chlorophyll metabolism - Bombyx mori (domestic silkworm)
00980 Metabolism of xenobiotics by cytochrome P450 - Bombyx mori (domestic silkworm)
00982 Drug metabolism - cytochrome P450 - Bombyx mori (domestic silkworm)
00983 Drug metabolism - other enzymes - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
00053 Ascorbate and aldarate metabolism - Bombyx mori (domestic silkworm)
00830 Retinol metabolism - Bombyx mori (domestic silkworm)
00860 Porphyrin and chlorophyll metabolism - Bombyx mori (domestic silkworm)
00980 Metabolism of xenobiotics by cytochrome P450 - Bombyx mori (domestic silkworm)
00982 Drug metabolism - cytochrome P450 - Bombyx mori (domestic silkworm)
00983 Drug metabolism - other enzymes - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
GO
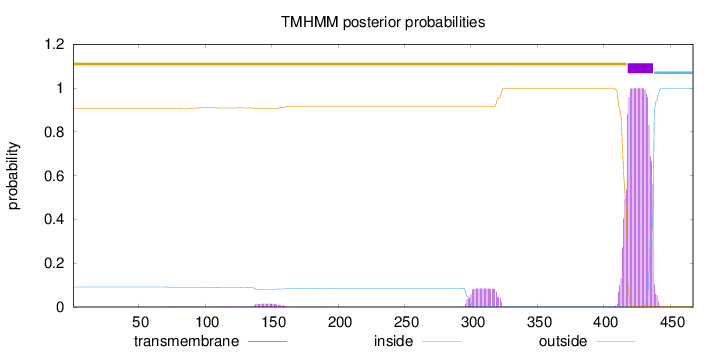
Topology
Subcellular location
Membrane
Length:
467
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.05577
Exp number, first 60 AAs:
0.00042
Total prob of N-in:
0.09185
outside
1 - 417
TMhelix
418 - 437
inside
438 - 467
Population Genetic Test Statistics
Pi
230.942903
Theta
158.475064
Tajima's D
0.927218
CLR
0.023356
CSRT
0.64276786160692
Interpretation
Uncertain
