Gene
KWMTBOMO10968
Pre Gene Modal
BGIBMGA008508
Annotation
UDP-glycosyltransferase_UGT43B1_precursor_[Bombyx_mori]
Full name
UDP-glucuronosyltransferase
Location in the cell
PlasmaMembrane Reliability : 2.113
Sequence
CDS
ATGAAAAACATATCGAAATTAAGACAAATACCACTCGAGTACAAAGTGAGCGGAAGCGATGTTGTTGATATAGACGATACTCTAAAAAACCTTCCTGGTATATTGGTTAATTTTCATAAAGCTTTGGACACAGCGAGAGCTTTCAAGAATTTAGCTAACTCAAATTGTAATAAATTGATGTCCAACAAAGAAATACAAGGAATAATATCGTCGAAGACTAAATTTGATTTAGTGATAGTTGAGCAATTTGTGACGGACTGCGGTCTTGCTGTTGCCTTTAAGTTGAATGCACCCATTGTCGGTATAACTGCTCATATTTTGATGCCGTGGACTTATTCAAGGTTGGGTGCGCTCAATCACCCGGCATACGTTCCGAATCATTTCATTGGTAGCGGAACCAAGCCTGGCTTTTGGGATAAGATACAAAGTGCACTAATTAATATCGCTTTCAACATTTACTTTAAGTATGTAATTCAAAAAAGCGACCAAATGATAATCAATAGTGTTTTTGAAGATGTACCGGACTTGGATGAGATCGGAAAGAATATTTCATTGATTTTATTGAATCAGTATTTTCCTTTGACTGGATCTCGGTTATATGGAGCGAATGTTATTGAAGTTGGTGGTTTACATATCAAAGAAAATACAACTATAGATGATGAGGAAATTAAAAGCTTCATTGACAAGGCAGAAAGCGACGTCATTTACATTAGTTTTGGAACTGTTGCATCTAATTTTCCGGATAGAATAATTAAAGAAATAATAAATTTTATCACGAAAAGTTCCGTAAAGGTTCTATGGAAAATTGACAATGTTGGTAATTTGAATTTGCCGAAGAATGTTTTAATAAGAAAATGGTTTCCGCAGACTGCTGTTTTGTGTCATCCAAAAGTGAAAGCATTTATCACACATTCTGGAATGTTAAGTTCAATTGAAGCTATGCATTGTGGGGTGCCAGTCATAAGCGTTCCGTTGTTCGGAGACCAATTCGCGAACGCAGCGGCAGCTACTGAAATTGGCTTGGGAGTTACCATTGATGTTTCCACAATGAATGAAAGAAAAATCAATCAAGCCCTAAAAACTGTGATGCAAGATTCTTATCAGATAAGGGCTCAAAATTTGTCTGCTTTGTGGAGAGATAGGCCCGTGTCACCTCTGAATTTAGCTATATTTTGGATTGAATACGTGATTCGGCACAAAGAAGACGACATATAA
Protein
MKNISKLRQIPLEYKVSGSDVVDIDDTLKNLPGILVNFHKALDTARAFKNLANSNCNKLMSNKEIQGIISSKTKFDLVIVEQFVTDCGLAVAFKLNAPIVGITAHILMPWTYSRLGALNHPAYVPNHFIGSGTKPGFWDKIQSALINIAFNIYFKYVIQKSDQMIINSVFEDVPDLDEIGKNISLILLNQYFPLTGSRLYGANVIEVGGLHIKENTTIDDEEIKSFIDKAESDVIYISFGTVASNFPDRIIKEIINFITKSSVKVLWKIDNVGNLNLPKNVLIRKWFPQTAVLCHPKVKAFITHSGMLSSIEAMHCGVPVISVPLFGDQFANAAAATEIGLGVTIDVSTMNERKINQALKTVMQDSYQIRAQNLSALWRDRPVSPLNLAIFWIEYVIRHKEDDI
Summary
Catalytic Activity
glucuronate acceptor + UDP-alpha-D-glucuronate = acceptor beta-D-glucuronoside + H(+) + UDP
Similarity
Belongs to the UDP-glycosyltransferase family.
Feature
chain UDP-glucuronosyltransferase
Uniprot
G9LPV8
H9JG61
A0A2A4JYW3
A0A191T1N2
A0A2H1VZI1
A0A2A4JZH0
+ More
G9LPR4 A0A0L7LQI5 A0A191T2H3 A0A2H4WB66 A0A1L8D6L6 A0A3S2LHF3 A0A2A4JZV9 A0A024AHN3 A0A2H4WB56 A0A212EWE0 G9LPR2 A0A2H1W091 G9LPV6 A0A191T1P6 A0A286MXP4 A0A194PHJ2 A0A1L8D6F4 A0A2H4WB57 G9LPV7 I4DM95 G9LPR3 G9LPV5 A0A212EWH2 A0A194RRD0 H9JYK2 A0A194PIG8 A0A194RRL0 A0A0C9RNU1 D2A2V5 A0A194Q839 A0A194RQ88 A0A182NE44 A0A2P8ZG78 A0A1L8E1G2 A0A182PJ64 A0A2M3Z1Z8 A0A084VP43 A0A1J1HE03 A0A2M3Z1Z1 A0A182GI26 A0A3S2PBM0 V5H2B2 A0A1S4F2K0 Q17HG0 A0A1I8N2C0 A0A336LZ19 A0A182V6Z3 A0A336KDN1 A0A336LVC8 A0A1Y1LIT5 T1PFI1 A0A182IX39 A0A182W9Q0 A0A2P8ZG70 B0WBP4 A0A182F4M1 A0A2A4JHL4 A0A182SC34 A0A1Q3FPW9 A0A182UEM4 A0A3Q0JH20 A0A182RBM8 A0A1Q3FPY1 A0A182YGV2 A0A139WIW7 A0A3Q0JLK0 Q17NF9 G9LPR5 A0A1W7R8J2 A0A182WT93 A0A1S4GXF8 W5J9T2 A0A212EWF8 A0A2J7R4V3 A0A182F4L8 A0A1I8NWG2 A0A182HV86 A0A1W4WYE4 A0A2J7R4V1 A0A286MXP5 B3NL93 W8AR85 S4PCR1 A0A067QGQ4 A0A1W4WMF1 A0A1I8NWF4 A0A2J7R4S8 A0A1S4EWK0 A0A2H4WB74 A0A1B6DGT0 A0A0K8UGE5 A0A0K8UM10 A0A2J7QFA8 A0A2J7R4W8
G9LPR4 A0A0L7LQI5 A0A191T2H3 A0A2H4WB66 A0A1L8D6L6 A0A3S2LHF3 A0A2A4JZV9 A0A024AHN3 A0A2H4WB56 A0A212EWE0 G9LPR2 A0A2H1W091 G9LPV6 A0A191T1P6 A0A286MXP4 A0A194PHJ2 A0A1L8D6F4 A0A2H4WB57 G9LPV7 I4DM95 G9LPR3 G9LPV5 A0A212EWH2 A0A194RRD0 H9JYK2 A0A194PIG8 A0A194RRL0 A0A0C9RNU1 D2A2V5 A0A194Q839 A0A194RQ88 A0A182NE44 A0A2P8ZG78 A0A1L8E1G2 A0A182PJ64 A0A2M3Z1Z8 A0A084VP43 A0A1J1HE03 A0A2M3Z1Z1 A0A182GI26 A0A3S2PBM0 V5H2B2 A0A1S4F2K0 Q17HG0 A0A1I8N2C0 A0A336LZ19 A0A182V6Z3 A0A336KDN1 A0A336LVC8 A0A1Y1LIT5 T1PFI1 A0A182IX39 A0A182W9Q0 A0A2P8ZG70 B0WBP4 A0A182F4M1 A0A2A4JHL4 A0A182SC34 A0A1Q3FPW9 A0A182UEM4 A0A3Q0JH20 A0A182RBM8 A0A1Q3FPY1 A0A182YGV2 A0A139WIW7 A0A3Q0JLK0 Q17NF9 G9LPR5 A0A1W7R8J2 A0A182WT93 A0A1S4GXF8 W5J9T2 A0A212EWF8 A0A2J7R4V3 A0A182F4L8 A0A1I8NWG2 A0A182HV86 A0A1W4WYE4 A0A2J7R4V1 A0A286MXP5 B3NL93 W8AR85 S4PCR1 A0A067QGQ4 A0A1W4WMF1 A0A1I8NWF4 A0A2J7R4S8 A0A1S4EWK0 A0A2H4WB74 A0A1B6DGT0 A0A0K8UGE5 A0A0K8UM10 A0A2J7QFA8 A0A2J7R4W8
EC Number
2.4.1.17
Pubmed
EMBL
JQ070265
AEW43181.1
BABH01002044
NWSH01000347
PCG77205.1
KU680307
+ More
ANI22013.1 ODYU01005418 SOQ46241.1 PCG77209.1 JQ070221 AEW43137.1 JTDY01000310 KOB77758.1 KU680305 ANI22011.1 KY202934 AUC64278.1 GEYN01000094 JAV02035.1 RSAL01000119 RVE46732.1 PCG77184.1 KF777118 AHY99689.1 KY202929 AUC64273.1 AGBW02012000 OWR45818.1 JQ070219 AEW43135.1 SOQ46242.1 JQ070263 AEW43179.1 KU680306 ANI22012.1 MF684362 ASX93996.1 KQ459604 KPI92518.1 GEYN01000096 JAV02033.1 KY202930 AUC64274.1 JQ070264 AEW43180.1 AK402413 BAM19035.1 JQ070220 AEW43136.1 JQ070262 AEW43178.1 OWR45817.1 KQ459765 KPJ20032.1 BABH01043998 KPI92519.1 KPJ20034.1 GBYB01008756 JAG78523.1 KQ971338 EFA02977.2 KQ459302 KPJ01707.1 KPJ20033.1 PYGN01000068 PSN55501.1 GFDF01001547 JAV12537.1 GGFM01001806 MBW22557.1 ATLV01014974 KE524999 KFB39737.1 CVRI01000001 CRK86175.1 GGFM01001770 MBW22521.1 JXUM01065169 KQ562333 KXJ76125.1 RVE46733.1 GALX01001513 JAB66953.1 CH477249 EAT46102.1 UFQS01000219 UFQT01000219 SSX01523.1 SSX21903.1 SSX01525.1 SSX21905.1 SSX01524.1 SSX21904.1 GEZM01054555 JAV73559.1 KA647552 AFP62181.1 PSN55494.1 DS231880 EDS42579.1 NWSH01001552 PCG70882.1 GFDL01005396 JAV29649.1 GFDL01005384 JAV29661.1 KYB27856.1 CH477199 EAT48259.1 JQ070222 AEW43138.1 GEHC01000181 JAV47464.1 AAAB01008964 ADMH02001962 ETN60168.1 OWR45816.1 NEVH01007402 PNF35864.1 APCN01002501 PNF35863.1 MF684363 ASX93997.1 CH954179 EDV54743.1 GAMC01019493 GAMC01019492 GAMC01019491 JAB87063.1 GAIX01004036 JAA88524.1 KK853490 KDR07235.1 PNF35836.1 KY202936 AUC64280.1 GEDC01012446 JAS24852.1 GDHF01026592 JAI25722.1 GDHF01024708 JAI27606.1 NEVH01015300 PNF27276.1 PNF35870.1
ANI22013.1 ODYU01005418 SOQ46241.1 PCG77209.1 JQ070221 AEW43137.1 JTDY01000310 KOB77758.1 KU680305 ANI22011.1 KY202934 AUC64278.1 GEYN01000094 JAV02035.1 RSAL01000119 RVE46732.1 PCG77184.1 KF777118 AHY99689.1 KY202929 AUC64273.1 AGBW02012000 OWR45818.1 JQ070219 AEW43135.1 SOQ46242.1 JQ070263 AEW43179.1 KU680306 ANI22012.1 MF684362 ASX93996.1 KQ459604 KPI92518.1 GEYN01000096 JAV02033.1 KY202930 AUC64274.1 JQ070264 AEW43180.1 AK402413 BAM19035.1 JQ070220 AEW43136.1 JQ070262 AEW43178.1 OWR45817.1 KQ459765 KPJ20032.1 BABH01043998 KPI92519.1 KPJ20034.1 GBYB01008756 JAG78523.1 KQ971338 EFA02977.2 KQ459302 KPJ01707.1 KPJ20033.1 PYGN01000068 PSN55501.1 GFDF01001547 JAV12537.1 GGFM01001806 MBW22557.1 ATLV01014974 KE524999 KFB39737.1 CVRI01000001 CRK86175.1 GGFM01001770 MBW22521.1 JXUM01065169 KQ562333 KXJ76125.1 RVE46733.1 GALX01001513 JAB66953.1 CH477249 EAT46102.1 UFQS01000219 UFQT01000219 SSX01523.1 SSX21903.1 SSX01525.1 SSX21905.1 SSX01524.1 SSX21904.1 GEZM01054555 JAV73559.1 KA647552 AFP62181.1 PSN55494.1 DS231880 EDS42579.1 NWSH01001552 PCG70882.1 GFDL01005396 JAV29649.1 GFDL01005384 JAV29661.1 KYB27856.1 CH477199 EAT48259.1 JQ070222 AEW43138.1 GEHC01000181 JAV47464.1 AAAB01008964 ADMH02001962 ETN60168.1 OWR45816.1 NEVH01007402 PNF35864.1 APCN01002501 PNF35863.1 MF684363 ASX93997.1 CH954179 EDV54743.1 GAMC01019493 GAMC01019492 GAMC01019491 JAB87063.1 GAIX01004036 JAA88524.1 KK853490 KDR07235.1 PNF35836.1 KY202936 AUC64280.1 GEDC01012446 JAS24852.1 GDHF01026592 JAI25722.1 GDHF01024708 JAI27606.1 NEVH01015300 PNF27276.1 PNF35870.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000283053
UP000007151
UP000053268
+ More
UP000053240 UP000007266 UP000075884 UP000245037 UP000075885 UP000030765 UP000183832 UP000069940 UP000249989 UP000008820 UP000095301 UP000075903 UP000075880 UP000075920 UP000002320 UP000069272 UP000075901 UP000075902 UP000079169 UP000075900 UP000076408 UP000076407 UP000000673 UP000235965 UP000095300 UP000075840 UP000192223 UP000008711 UP000027135
UP000053240 UP000007266 UP000075884 UP000245037 UP000075885 UP000030765 UP000183832 UP000069940 UP000249989 UP000008820 UP000095301 UP000075903 UP000075880 UP000075920 UP000002320 UP000069272 UP000075901 UP000075902 UP000079169 UP000075900 UP000076408 UP000076407 UP000000673 UP000235965 UP000095300 UP000075840 UP000192223 UP000008711 UP000027135
Pfam
Interpro
IPR002213
UDP_glucos_trans
+ More
IPR035595 UDP_glycos_trans_CS
IPR001041 2Fe-2S_ferredoxin-type
IPR036010 2Fe-2S_ferredoxin-like_sf
IPR022533 Cox20
IPR036683 CO_DH_flav_C_dom_sf
IPR036856 Ald_Oxase/Xan_DH_a/b_sf
IPR006058 2Fe2S_fd_BS
IPR016166 FAD-bd_PCMH
IPR002346 Mopterin_DH_FAD-bd
IPR037165 AldOxase/xan_DH_Mopterin-bd_sf
IPR000674 Ald_Oxase/Xan_DH_a/b
IPR002888 2Fe-2S-bd
IPR014307 Xanthine_DH_ssu
IPR036884 2Fe-2S-bd_dom_sf
IPR005107 CO_DH_flav_C
IPR008274 AldOxase/xan_DH_Mopterin-bd
IPR017978 GPCR_3_C
IPR036318 FAD-bd_PCMH-like_sf
IPR035595 UDP_glycos_trans_CS
IPR001041 2Fe-2S_ferredoxin-type
IPR036010 2Fe-2S_ferredoxin-like_sf
IPR022533 Cox20
IPR036683 CO_DH_flav_C_dom_sf
IPR036856 Ald_Oxase/Xan_DH_a/b_sf
IPR006058 2Fe2S_fd_BS
IPR016166 FAD-bd_PCMH
IPR002346 Mopterin_DH_FAD-bd
IPR037165 AldOxase/xan_DH_Mopterin-bd_sf
IPR000674 Ald_Oxase/Xan_DH_a/b
IPR002888 2Fe-2S-bd
IPR014307 Xanthine_DH_ssu
IPR036884 2Fe-2S-bd_dom_sf
IPR005107 CO_DH_flav_C
IPR008274 AldOxase/xan_DH_Mopterin-bd
IPR017978 GPCR_3_C
IPR036318 FAD-bd_PCMH-like_sf
SUPFAM
CDD
ProteinModelPortal
G9LPV8
H9JG61
A0A2A4JYW3
A0A191T1N2
A0A2H1VZI1
A0A2A4JZH0
+ More
G9LPR4 A0A0L7LQI5 A0A191T2H3 A0A2H4WB66 A0A1L8D6L6 A0A3S2LHF3 A0A2A4JZV9 A0A024AHN3 A0A2H4WB56 A0A212EWE0 G9LPR2 A0A2H1W091 G9LPV6 A0A191T1P6 A0A286MXP4 A0A194PHJ2 A0A1L8D6F4 A0A2H4WB57 G9LPV7 I4DM95 G9LPR3 G9LPV5 A0A212EWH2 A0A194RRD0 H9JYK2 A0A194PIG8 A0A194RRL0 A0A0C9RNU1 D2A2V5 A0A194Q839 A0A194RQ88 A0A182NE44 A0A2P8ZG78 A0A1L8E1G2 A0A182PJ64 A0A2M3Z1Z8 A0A084VP43 A0A1J1HE03 A0A2M3Z1Z1 A0A182GI26 A0A3S2PBM0 V5H2B2 A0A1S4F2K0 Q17HG0 A0A1I8N2C0 A0A336LZ19 A0A182V6Z3 A0A336KDN1 A0A336LVC8 A0A1Y1LIT5 T1PFI1 A0A182IX39 A0A182W9Q0 A0A2P8ZG70 B0WBP4 A0A182F4M1 A0A2A4JHL4 A0A182SC34 A0A1Q3FPW9 A0A182UEM4 A0A3Q0JH20 A0A182RBM8 A0A1Q3FPY1 A0A182YGV2 A0A139WIW7 A0A3Q0JLK0 Q17NF9 G9LPR5 A0A1W7R8J2 A0A182WT93 A0A1S4GXF8 W5J9T2 A0A212EWF8 A0A2J7R4V3 A0A182F4L8 A0A1I8NWG2 A0A182HV86 A0A1W4WYE4 A0A2J7R4V1 A0A286MXP5 B3NL93 W8AR85 S4PCR1 A0A067QGQ4 A0A1W4WMF1 A0A1I8NWF4 A0A2J7R4S8 A0A1S4EWK0 A0A2H4WB74 A0A1B6DGT0 A0A0K8UGE5 A0A0K8UM10 A0A2J7QFA8 A0A2J7R4W8
G9LPR4 A0A0L7LQI5 A0A191T2H3 A0A2H4WB66 A0A1L8D6L6 A0A3S2LHF3 A0A2A4JZV9 A0A024AHN3 A0A2H4WB56 A0A212EWE0 G9LPR2 A0A2H1W091 G9LPV6 A0A191T1P6 A0A286MXP4 A0A194PHJ2 A0A1L8D6F4 A0A2H4WB57 G9LPV7 I4DM95 G9LPR3 G9LPV5 A0A212EWH2 A0A194RRD0 H9JYK2 A0A194PIG8 A0A194RRL0 A0A0C9RNU1 D2A2V5 A0A194Q839 A0A194RQ88 A0A182NE44 A0A2P8ZG78 A0A1L8E1G2 A0A182PJ64 A0A2M3Z1Z8 A0A084VP43 A0A1J1HE03 A0A2M3Z1Z1 A0A182GI26 A0A3S2PBM0 V5H2B2 A0A1S4F2K0 Q17HG0 A0A1I8N2C0 A0A336LZ19 A0A182V6Z3 A0A336KDN1 A0A336LVC8 A0A1Y1LIT5 T1PFI1 A0A182IX39 A0A182W9Q0 A0A2P8ZG70 B0WBP4 A0A182F4M1 A0A2A4JHL4 A0A182SC34 A0A1Q3FPW9 A0A182UEM4 A0A3Q0JH20 A0A182RBM8 A0A1Q3FPY1 A0A182YGV2 A0A139WIW7 A0A3Q0JLK0 Q17NF9 G9LPR5 A0A1W7R8J2 A0A182WT93 A0A1S4GXF8 W5J9T2 A0A212EWF8 A0A2J7R4V3 A0A182F4L8 A0A1I8NWG2 A0A182HV86 A0A1W4WYE4 A0A2J7R4V1 A0A286MXP5 B3NL93 W8AR85 S4PCR1 A0A067QGQ4 A0A1W4WMF1 A0A1I8NWF4 A0A2J7R4S8 A0A1S4EWK0 A0A2H4WB74 A0A1B6DGT0 A0A0K8UGE5 A0A0K8UM10 A0A2J7QFA8 A0A2J7R4W8
PDB
2O6L
E-value=5.26078e-20,
Score=241
Ontologies
PATHWAY
00040
Pentose and glucuronate interconversions - Bombyx mori (domestic silkworm)
00053 Ascorbate and aldarate metabolism - Bombyx mori (domestic silkworm)
00830 Retinol metabolism - Bombyx mori (domestic silkworm)
00860 Porphyrin and chlorophyll metabolism - Bombyx mori (domestic silkworm)
00980 Metabolism of xenobiotics by cytochrome P450 - Bombyx mori (domestic silkworm)
00982 Drug metabolism - cytochrome P450 - Bombyx mori (domestic silkworm)
00983 Drug metabolism - other enzymes - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
00053 Ascorbate and aldarate metabolism - Bombyx mori (domestic silkworm)
00830 Retinol metabolism - Bombyx mori (domestic silkworm)
00860 Porphyrin and chlorophyll metabolism - Bombyx mori (domestic silkworm)
00980 Metabolism of xenobiotics by cytochrome P450 - Bombyx mori (domestic silkworm)
00982 Drug metabolism - cytochrome P450 - Bombyx mori (domestic silkworm)
00983 Drug metabolism - other enzymes - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
GO
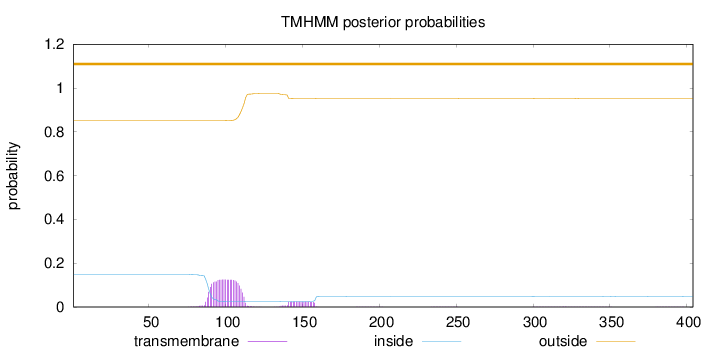
Topology
Subcellular location
Membrane
Length:
404
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.27128
Exp number, first 60 AAs:
0
Total prob of N-in:
0.14939
outside
1 - 404
Population Genetic Test Statistics
Pi
219.683867
Theta
128.578852
Tajima's D
2.424756
CLR
0.166154
CSRT
0.935453227338633
Interpretation
Uncertain
