Gene
KWMTBOMO10966 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008507
Annotation
mitochondrial_import_inner_membrane_translocase_subunit_Tim9A_[Bombyx_mori]
Full name
Mitochondrial import inner membrane translocase subunit Tim9
Location in the cell
Cytoplasmic Reliability : 1.123 Extracellular Reliability : 1.577 Nuclear Reliability : 1.444
Sequence
CDS
ATGGCGACCCCTGAAATGATGAGTGAAGTTGATCAAATTAAAACTTTCAAGGATTTCCTTGTACAATACAACAAGCTGTCTGAACTTTGTTTCAATGATTGCATTCATGATTTTACGTCGAGGACTTTAAAATCTACCGAGGACAAATGCACTGTCAACTGTATGGAGAAATATCTACGAATGAATCAGAGGGTTTCACAAAGATTCCATGAGTTCCAAATGCTTGCCAATGAAAACATGTTAGCATTAGCACAAAAATCTGGTAACCCTAGTTAA
Protein
MATPEMMSEVDQIKTFKDFLVQYNKLSELCFNDCIHDFTSRTLKSTEDKCTVNCMEKYLRMNQRVSQRFHEFQMLANENMLALAQKSGNPS
Summary
Description
Mitochondrial intermembrane chaperone that participates in the import and insertion of multi-pass transmembrane proteins into the mitochondrial inner membrane. May also be required for the transfer of beta-barrel precursors from the TOM complex to the sorting and assembly machinery (SAM complex) of the outer membrane. Acts as a chaperone-like protein that protects the hydrophobic precursors from aggregation and guide them through the mitochondrial intermembrane space (By similarity).
Subunit
Heterohexamer; composed of 3 copies of Tim9 and 3 copies of Tim10, named soluble 70 kDa complex. The complex associates with the Tim22 component of the TIM22 complex. Interacts with multi-pass transmembrane proteins in transit (By similarity).
Similarity
Belongs to the small Tim family.
Keywords
Chaperone
Complete proteome
Disulfide bond
Membrane
Metal-binding
Mitochondrion
Mitochondrion inner membrane
Protein transport
Reference proteome
Translocation
Transport
Zinc
Feature
chain Mitochondrial import inner membrane translocase subunit Tim9
Uniprot
Q1HQ37
A0A0L7LQT2
A0A194RV37
A0A194PGU0
A0A3S2TI84
A0A2A4JYW1
+ More
A0A212EWG6 A0A2H1VZH6 H9JG60 A0A0M4EVE4 B4Q2B1 B3NWP1 A0A0K8TNY6 A0A1W4V7X0 B4JLP9 B3MXN9 A0A0L0BRG1 B4MEI0 B4L1S4 B4IGS1 D2A0D3 A0A1I8MNV7 A0A1I8Q436 B4R4C1 X2JEU5 Q9VYD7 A0A3B0K5B6 A0A1J1IB76 A0A1B0D8V3 A0A2R5LE88 A0A1B0ANA2 A0A1A9ZIA6 E2C356 Q7QE81 A0A182I9L0 A0A182PM78 A0A1A9XH46 T1DU80 E0VM34 A0A2M3ZIY7 A0A2M4AS49 W5J825 A0A182FRL1 A0A182KFQ1 A0A2M4C4U0 A0A2P6KR19 A0A0P4VNJ0 R4FJR0 A0A087U9H9 A0A182RJ33 A0A182VVC8 A0A182XY51 A0A182URN0 A0A1L8E114 A0A182X5C7 B4NEH4 A0A182N0D1 A0A1W4XM17 A0A195F992 B5DIU5 B4G7R1 D6QYC4 A0A182QS37 B4H2W4 G3MQU9 A0A182LIK4 A0A2R7WK63 A0A0C9RRK7 A0A1E1WZ18 A0A023FVX8 A0A023FFZ5 A0A195BYD8 A0A0P5TQ71 A0A0N8BWM7 A0A151IR04 B7PDS2 B0XGR7 A0A151JM11 A0A2R7VRX3 A0A1V9XW16 B7QBV2 A0A0K8R660 A0A1Q3F993 N6TWT6 E1ZWB8 E9GYJ3 F4WX31 A0A151WMY7 T1H778 A0A0N7ZPC9 A0A1B6DMF3 A0A0P4ZBJ4 A0A131XPK3 A0A154NZL8 N6TLJ8 A0A067RT82 A0A0P5V377 D6QYC2 A0A232FBV4 K7IPB3
A0A212EWG6 A0A2H1VZH6 H9JG60 A0A0M4EVE4 B4Q2B1 B3NWP1 A0A0K8TNY6 A0A1W4V7X0 B4JLP9 B3MXN9 A0A0L0BRG1 B4MEI0 B4L1S4 B4IGS1 D2A0D3 A0A1I8MNV7 A0A1I8Q436 B4R4C1 X2JEU5 Q9VYD7 A0A3B0K5B6 A0A1J1IB76 A0A1B0D8V3 A0A2R5LE88 A0A1B0ANA2 A0A1A9ZIA6 E2C356 Q7QE81 A0A182I9L0 A0A182PM78 A0A1A9XH46 T1DU80 E0VM34 A0A2M3ZIY7 A0A2M4AS49 W5J825 A0A182FRL1 A0A182KFQ1 A0A2M4C4U0 A0A2P6KR19 A0A0P4VNJ0 R4FJR0 A0A087U9H9 A0A182RJ33 A0A182VVC8 A0A182XY51 A0A182URN0 A0A1L8E114 A0A182X5C7 B4NEH4 A0A182N0D1 A0A1W4XM17 A0A195F992 B5DIU5 B4G7R1 D6QYC4 A0A182QS37 B4H2W4 G3MQU9 A0A182LIK4 A0A2R7WK63 A0A0C9RRK7 A0A1E1WZ18 A0A023FVX8 A0A023FFZ5 A0A195BYD8 A0A0P5TQ71 A0A0N8BWM7 A0A151IR04 B7PDS2 B0XGR7 A0A151JM11 A0A2R7VRX3 A0A1V9XW16 B7QBV2 A0A0K8R660 A0A1Q3F993 N6TWT6 E1ZWB8 E9GYJ3 F4WX31 A0A151WMY7 T1H778 A0A0N7ZPC9 A0A1B6DMF3 A0A0P4ZBJ4 A0A131XPK3 A0A154NZL8 N6TLJ8 A0A067RT82 A0A0P5V377 D6QYC2 A0A232FBV4 K7IPB3
Pubmed
26227816
26354079
22118469
19121390
17994087
17550304
+ More
26369729 26108605 18362917 19820115 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20798317 12364791 14747013 17210077 20566863 20920257 23761445 27129103 25244985 15632085 20351054 22216098 20966253 26131772 28503490 28327890 23537049 21292972 21719571 28049606 24845553 28648823 20075255
26369729 26108605 18362917 19820115 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20798317 12364791 14747013 17210077 20566863 20920257 23761445 27129103 25244985 15632085 20351054 22216098 20966253 26131772 28503490 28327890 23537049 21292972 21719571 28049606 24845553 28648823 20075255
EMBL
DQ443215
ABF51304.1
JTDY01000310
KOB77759.1
KQ459765
KPJ20031.1
+ More
KQ459604 KPI92517.1 RSAL01000119 RVE46730.1 NWSH01000347 PCG77211.1 AGBW02012000 AGBW02009830 OWR45819.1 OWR49796.1 ODYU01005418 SOQ46239.1 BABH01002044 CP012528 ALC49926.1 CM000162 EDX01572.1 CH954180 EDV47203.1 GDAI01001747 JAI15856.1 CH916371 EDV91660.1 CH902630 EDV38504.1 JRES01001467 KNC22665.1 CH940664 EDW62955.1 CH933810 EDW06727.1 CH480836 EDW49021.1 KQ971338 EFA02499.1 CM000366 EDX17823.1 AE014298 KX531321 AHN59678.1 ANY27131.1 BT025812 OUUW01000003 SPP78658.1 CVRI01000044 CRK96812.1 AJVK01004114 GGLE01003705 MBY07831.1 JXJN01000772 GL452292 EFN77588.1 AAAB01008847 EAA06794.4 APCN01000999 GAMD01000389 JAB01202.1 DS235286 EEB14440.1 GGFM01007786 MBW28537.1 GGFK01010241 MBW43562.1 ADMH02002105 ETN59010.1 GGFJ01011173 MBW60314.1 MWRG01006682 PRD28775.1 GDKW01002673 JAI53922.1 ACPB03006777 GAHY01002173 JAA75337.1 KK118852 KFM74018.1 GFDF01001666 JAV12418.1 CH964239 EDW82143.1 KQ981727 KYN37008.1 CH379060 EDY70238.1 CH479180 EDW29332.1 HM017571 HM017572 HM017573 HM017574 HM017575 HM017576 HM017577 HM017578 HM017579 HM017580 HM017581 HM017582 ADG64944.1 ADG64945.1 ADG64946.1 ADG64947.1 ADG64948.1 ADG64949.1 ADG64950.1 ADG64951.1 ADG64952.1 ADG64953.1 ADG64954.1 ADG64955.1 AXCN02002175 CH479204 EDW30681.1 JO844250 AEO35867.1 KK854960 PTY19958.1 GBZX01002879 JAG89861.1 GFAC01007162 JAT92026.1 GBBL01002412 JAC24908.1 GBBK01004467 JAC20015.1 KQ976396 KYM92951.1 GDIP01128504 GDIQ01087219 LRGB01001763 JAL75210.1 JAN07518.1 KZS10673.1 GDIQ01137643 JAL14083.1 KQ976759 KYN08551.1 ABJB011105252 DS691488 EEC04744.1 DS233058 EDS27790.1 KQ978960 KYN26928.1 KK854033 PTY09898.1 MNPL01003396 OQR77548.1 ABJB010633092 DS903634 EEC16324.1 GADI01007440 JAA66368.1 GFDL01010956 JAV24089.1 APGK01057945 KB741283 KB631738 ENN70772.1 ERL85678.1 GL434791 EFN74522.1 GL732575 EFX75474.1 GL888417 EGI61274.1 KQ982934 KYQ49203.1 GDIP01225705 JAI97696.1 GEDC01010498 JAS26800.1 GDIP01228055 JAI95346.1 GEFH01000436 JAP68145.1 KQ434786 KZC05115.1 APGK01031540 KB740814 ENN78863.1 KK852478 KDR23029.1 GDIP01106210 JAL97504.1 HM017569 HM017570 ADG64942.1 ADG64943.1 NNAY01000447 OXU28316.1
KQ459604 KPI92517.1 RSAL01000119 RVE46730.1 NWSH01000347 PCG77211.1 AGBW02012000 AGBW02009830 OWR45819.1 OWR49796.1 ODYU01005418 SOQ46239.1 BABH01002044 CP012528 ALC49926.1 CM000162 EDX01572.1 CH954180 EDV47203.1 GDAI01001747 JAI15856.1 CH916371 EDV91660.1 CH902630 EDV38504.1 JRES01001467 KNC22665.1 CH940664 EDW62955.1 CH933810 EDW06727.1 CH480836 EDW49021.1 KQ971338 EFA02499.1 CM000366 EDX17823.1 AE014298 KX531321 AHN59678.1 ANY27131.1 BT025812 OUUW01000003 SPP78658.1 CVRI01000044 CRK96812.1 AJVK01004114 GGLE01003705 MBY07831.1 JXJN01000772 GL452292 EFN77588.1 AAAB01008847 EAA06794.4 APCN01000999 GAMD01000389 JAB01202.1 DS235286 EEB14440.1 GGFM01007786 MBW28537.1 GGFK01010241 MBW43562.1 ADMH02002105 ETN59010.1 GGFJ01011173 MBW60314.1 MWRG01006682 PRD28775.1 GDKW01002673 JAI53922.1 ACPB03006777 GAHY01002173 JAA75337.1 KK118852 KFM74018.1 GFDF01001666 JAV12418.1 CH964239 EDW82143.1 KQ981727 KYN37008.1 CH379060 EDY70238.1 CH479180 EDW29332.1 HM017571 HM017572 HM017573 HM017574 HM017575 HM017576 HM017577 HM017578 HM017579 HM017580 HM017581 HM017582 ADG64944.1 ADG64945.1 ADG64946.1 ADG64947.1 ADG64948.1 ADG64949.1 ADG64950.1 ADG64951.1 ADG64952.1 ADG64953.1 ADG64954.1 ADG64955.1 AXCN02002175 CH479204 EDW30681.1 JO844250 AEO35867.1 KK854960 PTY19958.1 GBZX01002879 JAG89861.1 GFAC01007162 JAT92026.1 GBBL01002412 JAC24908.1 GBBK01004467 JAC20015.1 KQ976396 KYM92951.1 GDIP01128504 GDIQ01087219 LRGB01001763 JAL75210.1 JAN07518.1 KZS10673.1 GDIQ01137643 JAL14083.1 KQ976759 KYN08551.1 ABJB011105252 DS691488 EEC04744.1 DS233058 EDS27790.1 KQ978960 KYN26928.1 KK854033 PTY09898.1 MNPL01003396 OQR77548.1 ABJB010633092 DS903634 EEC16324.1 GADI01007440 JAA66368.1 GFDL01010956 JAV24089.1 APGK01057945 KB741283 KB631738 ENN70772.1 ERL85678.1 GL434791 EFN74522.1 GL732575 EFX75474.1 GL888417 EGI61274.1 KQ982934 KYQ49203.1 GDIP01225705 JAI97696.1 GEDC01010498 JAS26800.1 GDIP01228055 JAI95346.1 GEFH01000436 JAP68145.1 KQ434786 KZC05115.1 APGK01031540 KB740814 ENN78863.1 KK852478 KDR23029.1 GDIP01106210 JAL97504.1 HM017569 HM017570 ADG64942.1 ADG64943.1 NNAY01000447 OXU28316.1
Proteomes
UP000037510
UP000053240
UP000053268
UP000283053
UP000218220
UP000007151
+ More
UP000005204 UP000092553 UP000002282 UP000008711 UP000192221 UP000001070 UP000007801 UP000037069 UP000008792 UP000009192 UP000001292 UP000007266 UP000095301 UP000095300 UP000000304 UP000000803 UP000268350 UP000183832 UP000092462 UP000092460 UP000092445 UP000008237 UP000007062 UP000075840 UP000075885 UP000092443 UP000009046 UP000000673 UP000069272 UP000075881 UP000015103 UP000054359 UP000075900 UP000075920 UP000076408 UP000075903 UP000076407 UP000007798 UP000075884 UP000192223 UP000078541 UP000001819 UP000008744 UP000075886 UP000075882 UP000078540 UP000076858 UP000078542 UP000001555 UP000002320 UP000078492 UP000192247 UP000019118 UP000030742 UP000000311 UP000000305 UP000007755 UP000075809 UP000015102 UP000076502 UP000027135 UP000215335 UP000002358
UP000005204 UP000092553 UP000002282 UP000008711 UP000192221 UP000001070 UP000007801 UP000037069 UP000008792 UP000009192 UP000001292 UP000007266 UP000095301 UP000095300 UP000000304 UP000000803 UP000268350 UP000183832 UP000092462 UP000092460 UP000092445 UP000008237 UP000007062 UP000075840 UP000075885 UP000092443 UP000009046 UP000000673 UP000069272 UP000075881 UP000015103 UP000054359 UP000075900 UP000075920 UP000076408 UP000075903 UP000076407 UP000007798 UP000075884 UP000192223 UP000078541 UP000001819 UP000008744 UP000075886 UP000075882 UP000078540 UP000076858 UP000078542 UP000001555 UP000002320 UP000078492 UP000192247 UP000019118 UP000030742 UP000000311 UP000000305 UP000007755 UP000075809 UP000015102 UP000076502 UP000027135 UP000215335 UP000002358
Pfam
PF02953 zf-Tim10_DDP
SUPFAM
SSF144122
SSF144122
Gene 3D
ProteinModelPortal
Q1HQ37
A0A0L7LQT2
A0A194RV37
A0A194PGU0
A0A3S2TI84
A0A2A4JYW1
+ More
A0A212EWG6 A0A2H1VZH6 H9JG60 A0A0M4EVE4 B4Q2B1 B3NWP1 A0A0K8TNY6 A0A1W4V7X0 B4JLP9 B3MXN9 A0A0L0BRG1 B4MEI0 B4L1S4 B4IGS1 D2A0D3 A0A1I8MNV7 A0A1I8Q436 B4R4C1 X2JEU5 Q9VYD7 A0A3B0K5B6 A0A1J1IB76 A0A1B0D8V3 A0A2R5LE88 A0A1B0ANA2 A0A1A9ZIA6 E2C356 Q7QE81 A0A182I9L0 A0A182PM78 A0A1A9XH46 T1DU80 E0VM34 A0A2M3ZIY7 A0A2M4AS49 W5J825 A0A182FRL1 A0A182KFQ1 A0A2M4C4U0 A0A2P6KR19 A0A0P4VNJ0 R4FJR0 A0A087U9H9 A0A182RJ33 A0A182VVC8 A0A182XY51 A0A182URN0 A0A1L8E114 A0A182X5C7 B4NEH4 A0A182N0D1 A0A1W4XM17 A0A195F992 B5DIU5 B4G7R1 D6QYC4 A0A182QS37 B4H2W4 G3MQU9 A0A182LIK4 A0A2R7WK63 A0A0C9RRK7 A0A1E1WZ18 A0A023FVX8 A0A023FFZ5 A0A195BYD8 A0A0P5TQ71 A0A0N8BWM7 A0A151IR04 B7PDS2 B0XGR7 A0A151JM11 A0A2R7VRX3 A0A1V9XW16 B7QBV2 A0A0K8R660 A0A1Q3F993 N6TWT6 E1ZWB8 E9GYJ3 F4WX31 A0A151WMY7 T1H778 A0A0N7ZPC9 A0A1B6DMF3 A0A0P4ZBJ4 A0A131XPK3 A0A154NZL8 N6TLJ8 A0A067RT82 A0A0P5V377 D6QYC2 A0A232FBV4 K7IPB3
A0A212EWG6 A0A2H1VZH6 H9JG60 A0A0M4EVE4 B4Q2B1 B3NWP1 A0A0K8TNY6 A0A1W4V7X0 B4JLP9 B3MXN9 A0A0L0BRG1 B4MEI0 B4L1S4 B4IGS1 D2A0D3 A0A1I8MNV7 A0A1I8Q436 B4R4C1 X2JEU5 Q9VYD7 A0A3B0K5B6 A0A1J1IB76 A0A1B0D8V3 A0A2R5LE88 A0A1B0ANA2 A0A1A9ZIA6 E2C356 Q7QE81 A0A182I9L0 A0A182PM78 A0A1A9XH46 T1DU80 E0VM34 A0A2M3ZIY7 A0A2M4AS49 W5J825 A0A182FRL1 A0A182KFQ1 A0A2M4C4U0 A0A2P6KR19 A0A0P4VNJ0 R4FJR0 A0A087U9H9 A0A182RJ33 A0A182VVC8 A0A182XY51 A0A182URN0 A0A1L8E114 A0A182X5C7 B4NEH4 A0A182N0D1 A0A1W4XM17 A0A195F992 B5DIU5 B4G7R1 D6QYC4 A0A182QS37 B4H2W4 G3MQU9 A0A182LIK4 A0A2R7WK63 A0A0C9RRK7 A0A1E1WZ18 A0A023FVX8 A0A023FFZ5 A0A195BYD8 A0A0P5TQ71 A0A0N8BWM7 A0A151IR04 B7PDS2 B0XGR7 A0A151JM11 A0A2R7VRX3 A0A1V9XW16 B7QBV2 A0A0K8R660 A0A1Q3F993 N6TWT6 E1ZWB8 E9GYJ3 F4WX31 A0A151WMY7 T1H778 A0A0N7ZPC9 A0A1B6DMF3 A0A0P4ZBJ4 A0A131XPK3 A0A154NZL8 N6TLJ8 A0A067RT82 A0A0P5V377 D6QYC2 A0A232FBV4 K7IPB3
PDB
2BSK
E-value=6.0884e-17,
Score=207
Ontologies
PANTHER
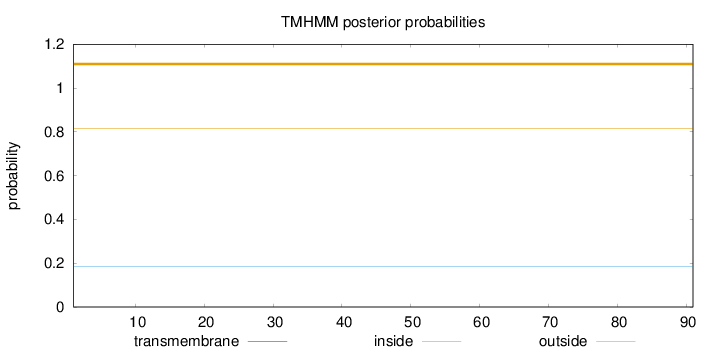
Topology
Subcellular location
Mitochondrion inner membrane
Length:
91
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00017
Exp number, first 60 AAs:
0
Total prob of N-in:
0.18343
outside
1 - 91
Population Genetic Test Statistics
Pi
268.583549
Theta
188.718962
Tajima's D
1.333877
CLR
0
CSRT
0.744862756862157
Interpretation
Uncertain
