Gene
KWMTBOMO10964
Pre Gene Modal
BGIBMGA008506
Annotation
PREDICTED:_serine/threonine-protein_kinase_PAK_mbt_[Papilio_polytes]
Full name
Non-specific serine/threonine protein kinase
Location in the cell
Mitochondrial Reliability : 1.837 Nuclear Reliability : 1.915
Sequence
CDS
ATGTTTTCTAAGAAGAAGAAAAAGCCACTTATTTCGCCGCCTAGTAATTTTGAACACAGAGTACACACTGGTTTCGATAAGCACGAGGGAAAGTTTGTTGGACTTCCTCTTCAGTGGGCTTCACTCGTTGGAAACAATCAGATATTAAAATCAACAAATCGACCTTTACCATTAGTGGATCCCTCTGAAATTACTCCTACAGAGATATTAGATTTGAAAACAATTGTTAGAGGTGATCACAGGATAACACCAGTGTCAGCTGTAGCAAGACTCTTGCCTGCAAACATTCCACAGCCACAACCAGCCAACCAACAAAATGGTTTTATATTGCCAAAGACATCAAATGTTGCTAGATCTAATTCTCTTAGAAGCACAAGTCCACCAAAAATTCGACGAGATTTACGTAATCAGGCTAATGTACCACCATCAGTACCCGAAGAATCTCCTCCTGTGCCTCCTGCTCCACAGCAGTATCCCAGTTTGAGACGGGAGCAGAGTAACTATGCTGTAAATAAGCCAATTCCAGAGTCTCCAGTGGAATGGTCACAGCATATGCACGAGGCAACAATCAGACAAGTTGCTGAAATTAATGATAACCATCATGCTGGTATGACTTATCAGAACATACAGAATGCGAACATGCCATATCAACATAGTCATCAAACACAAGCTGTTACTCACAATATGCATCTTGGCCTTAATGGAAACAACTTGCCCATTGGTGAGAACCAACAAGTTCTACGTCACCAGCATCCTGCATCGACCGCCGGTCCGGGCTCGACAGCGACCTCGATACCGCTCGCCGCATCAAAGCATGAATTGCGATTGACGCATGAACAGTTCCGCGCAGCGCTACGTCTAGTGGTGAGCGAGGGCGATCCCCGCGCTACGTTGACCGGCTTCAATAAGATCGGCAAGGGGAGCACGGGCATAGTTTGCTCGGCGACCGACACCCGCACGAGGAGAAGCGTCGCCGTCAAGATGATGAACTTGCGAAGGCAACAGAGACGAGAACTTCTGTTCAATGAAGTTGTGATAATGCGAGACTACCCGCATCCGAACATAGTGGAGATGTACGCTTCCTATCTCGTTGGTGACGAGCTATGGGTGGTCATGGAGTACATGGCGGGCGGCGCGCTCACCGACATCGTCACCCGCGTCCGGATGGACCCGGAACAGATCGCCACGGTCTGCAAGCAGTGCTTGAAGGCTCTGGCATTCCTTCACAGCCAAGGCGTTATACACAGAGACATCAAATCCGATTCAATACTAATGACGGCAGACGGGCGCGTCAAGTTATCAGACTTTGGCTTTTGTGCTCAAGTGTCGCAGGAATTACCTAAAAGAAAATCACTAGTTGGTACACCGTACTGGATGTCACCTGAGGTGATATCGAGGCTGCCGTACGGACCCGAGGTGGATGTGTGGTCGCTGGGCATTATGGTGGTCGAAATGGTAGACGGTGAGCCGCCATTCTTCAATGAACCTCCCCTACAGGCGATGCGTCGAATCCGCGACATGCCGGCGCCCCGGCCGCGCGGCGCCGCCCGCTGCCCGGCCGACCTGCTCGCGTTCGTGGAGAGCGCGCTGGTGCGGGACCCGGCGCTGCGGCTGTCCGCGCCGCGGCTGCTCTCGCACGCCTTCCTGCGCCGCGCCGGCCCGCCCGCGCTGCTCGTGCCGCTCATGCACGCGCCCTGA
Protein
MFSKKKKKPLISPPSNFEHRVHTGFDKHEGKFVGLPLQWASLVGNNQILKSTNRPLPLVDPSEITPTEILDLKTIVRGDHRITPVSAVARLLPANIPQPQPANQQNGFILPKTSNVARSNSLRSTSPPKIRRDLRNQANVPPSVPEESPPVPPAPQQYPSLRREQSNYAVNKPIPESPVEWSQHMHEATIRQVAEINDNHHAGMTYQNIQNANMPYQHSHQTQAVTHNMHLGLNGNNLPIGENQQVLRHQHPASTAGPGSTATSIPLAASKHELRLTHEQFRAALRLVVSEGDPRATLTGFNKIGKGSTGIVCSATDTRTRRSVAVKMMNLRRQQRRELLFNEVVIMRDYPHPNIVEMYASYLVGDELWVVMEYMAGGALTDIVTRVRMDPEQIATVCKQCLKALAFLHSQGVIHRDIKSDSILMTADGRVKLSDFGFCAQVSQELPKRKSLVGTPYWMSPEVISRLPYGPEVDVWSLGIMVVEMVDGEPPFFNEPPLQAMRRIRDMPAPRPRGAARCPADLLAFVESALVRDPALRLSAPRLLSHAFLRRAGPPALLVPLMHAP
Summary
Catalytic Activity
ATP + L-seryl-[protein] = ADP + H(+) + O-phospho-L-seryl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
Uniprot
EC Number
2.7.11.1
EMBL
BABH01002043
BABH01002044
NWSH01000347
PCG77214.1
ODYU01005418
SOQ46237.1
+ More
KQ459765 KPJ20043.1 KQ459604 KPI92529.1 AGBW02012905 OWR44232.1 GEZM01091144 JAV57081.1 GEDC01020557 GEDC01019933 JAS16741.1 JAS17365.1 GFDF01006997 JAV07087.1 ABLF02039472 ABLF02039487 GGMR01012025 MBY24644.1 GFXV01001299 MBW13104.1 JTDY01000310 KOB77757.1 KB202284 ESO90950.1
KQ459765 KPJ20043.1 KQ459604 KPI92529.1 AGBW02012905 OWR44232.1 GEZM01091144 JAV57081.1 GEDC01020557 GEDC01019933 JAS16741.1 JAS17365.1 GFDF01006997 JAV07087.1 ABLF02039472 ABLF02039487 GGMR01012025 MBY24644.1 GFXV01001299 MBW13104.1 JTDY01000310 KOB77757.1 KB202284 ESO90950.1
Proteomes
PRIDE
Interpro
SUPFAM
SSF56112
SSF56112
Gene 3D
ProteinModelPortal
PDB
2F57
E-value=4.3385e-116,
Score=1071
Ontologies
GO
PANTHER
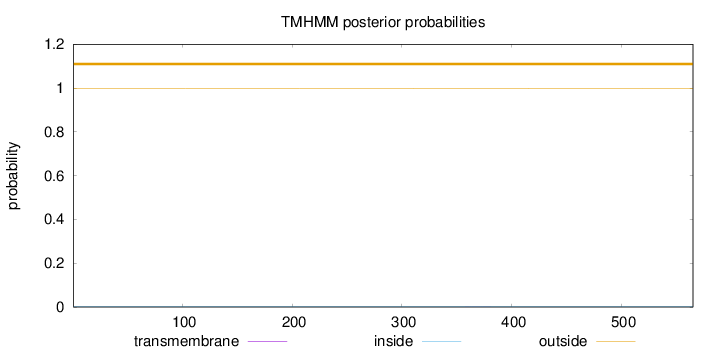
Topology
Length:
565
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00889999999999999
Exp number, first 60 AAs:
0.0013
Total prob of N-in:
0.00217
outside
1 - 565
Population Genetic Test Statistics
Pi
267.372059
Theta
198.002998
Tajima's D
1.087179
CLR
0.004898
CSRT
0.68201589920504
Interpretation
Uncertain
