Pre Gene Modal
BGIBMGA008505
Annotation
uncharacterized_protein_LOC101739460_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.425
Sequence
CDS
ATGACGAAAATAAATATTGAAGAAATTGACCTATATGCTGTGCTTGATTTACAAATAACTGCAACCGAATCGGAGATCAAAAAAGCATACAGAAAAAAGGCTTTACAATGCCACCCAGACAAAAACCCAGATGACCCTAAAGCGGCAGAAACATTTCACGAATTATCCCAAGCATTAGAAATATTAACTGATACTTCTGCTAGAGCAGCTTATGATAAAGTGCTACGTGCTAAAGCTTCCGCAAAGTTAAGGCATCAAGAATTAGACAGTAAAAGACAAAAACTCAAAGAAGATTTAGAAAAACGAGAAAGAGAAGCAGCATCTGGCACACATACAAACCTTACAGATTCTCAGAGACTTGCAAAAGAGATTGAAAGGCTACAAAGAGAGGGAAGTCGTCTATTATTGGAAGAGCAACAAAGAATGAAAAATGAAATACAAAAAAGTGTAGAACGAATGAAAGAACCTGTTTGGGATCCAAGTTTGAATAGAATTAAGATTAAGTGGAAAGTTGATAAAACTGATCAGAATAATAGTACTTATGATGAAGCTACATTAAGAAAGTTCTTGAAGAAATATGGTGACATTGTTGCTTTAGTAATATCATCCAAGAGTCGAGGTAGTGCCTTAATAGAATTTGCTACTAAGGAAGCAGCAGAAATGGCTGTTCAGTTAGAAAAAGGCCTACCTGACAACCCTTTAACTTTAAAATGGATAGAGATTATGAGTCTCTAG
Protein
MTKINIEEIDLYAVLDLQITATESEIKKAYRKKALQCHPDKNPDDPKAAETFHELSQALEILTDTSARAAYDKVLRAKASAKLRHQELDSKRQKLKEDLEKREREAASGTHTNLTDSQRLAKEIERLQREGSRLLLEEQQRMKNEIQKSVERMKEPVWDPSLNRIKIKWKVDKTDQNNSTYDEATLRKFLKKYGDIVALVISSKSRGSALIEFATKEAAEMAVQLEKGLPDNPLTLKWIEIMSL
Summary
Uniprot
H6VTQ5
A0A2A4JZL7
A0A0L7LUG6
A0A2H1VZH0
A0A212ERV4
S4NTV3
+ More
A0A194RRU4 A0A194PGL4 A0A0K8TM17 A0A182PU18 A0A182LAM0 A0A2M4BUY2 A0A182VY83 A0A182MG93 A0A182I8Q6 A0A182V816 A0A182WV32 A0A182XWN1 A0A2M4BVA5 A0A182JCN1 A0A182UAK4 A0A182F7V0 Q7QFV0 W5JHC6 A0A182QCL0 A0A023ELY6 A0A084VDX4 Q0C727 A0A336MZA4 A0A182RDB2 A0A182G8Y7 B0WLZ1 A0A336MG67 A0A182NSR1 A0A1A9VPH1 A0A1W4WP52 A0A336LH30 A0A1B0GCV5 A0A182JY39 A0A0L0BP01 N6UBY2 A0A2P8YRN5 W8CDG0 A0A2J7Q4T4 A0A182SNH9 B4KCZ6 A0A1I8M0Y4 A0A1Y1L7S7 T1PJB2 A0A087U683 A0A067R2E0 A0A0M4EQC9 Q293X1 B4LZI1 B4GLI1 D6WH30 A0A3B0KEF2 B4JUV2 A0A2R7WRY2 K7J968 A0A1J1HS15 B4HIC1 E0W330 B4QU60 A0A2J7Q4T1 A0A0K8SJS3 A0A0A9YUI0 A0A2R5LDQ6 A0A1B6DJR7 B3NZH1 B4PM33 A0A1I8QE65 A0A1W4ULR6 B4NBQ7 Q9VGR0 B3LZ51 A0A1B6GI15 A0A0K8VJW1 A0A3Q3GAV0 A0A034VVK0 A0A3Q4GFT0 A0A3B4AB67 R7U0E8 A0A154P3K1 A0A310SDC4 A0A0T6AYX5 Q4RLP2 H3DJA2 A0A1B6INP3 A0A0V0GAC0 R4FMM3 B9EQ16 A0A0N0BGU1 A0A3Q3LD35 A0A2Y9DT46 A0A287ALB8 A0A1U7UF69 A0A2I2YYS6 A0A1S3EMR0 A0A3Q3VNX4
A0A194RRU4 A0A194PGL4 A0A0K8TM17 A0A182PU18 A0A182LAM0 A0A2M4BUY2 A0A182VY83 A0A182MG93 A0A182I8Q6 A0A182V816 A0A182WV32 A0A182XWN1 A0A2M4BVA5 A0A182JCN1 A0A182UAK4 A0A182F7V0 Q7QFV0 W5JHC6 A0A182QCL0 A0A023ELY6 A0A084VDX4 Q0C727 A0A336MZA4 A0A182RDB2 A0A182G8Y7 B0WLZ1 A0A336MG67 A0A182NSR1 A0A1A9VPH1 A0A1W4WP52 A0A336LH30 A0A1B0GCV5 A0A182JY39 A0A0L0BP01 N6UBY2 A0A2P8YRN5 W8CDG0 A0A2J7Q4T4 A0A182SNH9 B4KCZ6 A0A1I8M0Y4 A0A1Y1L7S7 T1PJB2 A0A087U683 A0A067R2E0 A0A0M4EQC9 Q293X1 B4LZI1 B4GLI1 D6WH30 A0A3B0KEF2 B4JUV2 A0A2R7WRY2 K7J968 A0A1J1HS15 B4HIC1 E0W330 B4QU60 A0A2J7Q4T1 A0A0K8SJS3 A0A0A9YUI0 A0A2R5LDQ6 A0A1B6DJR7 B3NZH1 B4PM33 A0A1I8QE65 A0A1W4ULR6 B4NBQ7 Q9VGR0 B3LZ51 A0A1B6GI15 A0A0K8VJW1 A0A3Q3GAV0 A0A034VVK0 A0A3Q4GFT0 A0A3B4AB67 R7U0E8 A0A154P3K1 A0A310SDC4 A0A0T6AYX5 Q4RLP2 H3DJA2 A0A1B6INP3 A0A0V0GAC0 R4FMM3 B9EQ16 A0A0N0BGU1 A0A3Q3LD35 A0A2Y9DT46 A0A287ALB8 A0A1U7UF69 A0A2I2YYS6 A0A1S3EMR0 A0A3Q3VNX4
Pubmed
19121390
26434795
26227816
22118469
23622113
26354079
+ More
26369729 20966253 25244985 12364791 14747013 17210077 20920257 23761445 24945155 24438588 17510324 26483478 26108605 23537049 29403074 24495485 17994087 25315136 28004739 24845553 15632085 18362917 19820115 20075255 20566863 26823975 25401762 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 25186727 25463417 23254933 15496914 20433749 22398555
26369729 20966253 25244985 12364791 14747013 17210077 20920257 23761445 24945155 24438588 17510324 26483478 26108605 23537049 29403074 24495485 17994087 25315136 28004739 24845553 15632085 18362917 19820115 20075255 20566863 26823975 25401762 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 25186727 25463417 23254933 15496914 20433749 22398555
EMBL
BABH01002039
JN872890
AFC01229.1
NWSH01000347
PCG77216.1
JTDY01000072
+ More
KOB79029.1 ODYU01005418 SOQ46235.1 AGBW02012905 OWR44230.1 GAIX01012076 JAA80484.1 KQ459765 KPJ20045.1 KQ459604 KPI92531.1 GDAI01002628 JAI14975.1 GGFJ01007726 MBW56867.1 AXCM01007073 APCN01003107 GGFJ01007875 MBW57016.1 AAAB01008844 EAA06109.3 ADMH02001520 ETN62214.1 AXCN02000227 GAPW01003306 JAC10292.1 ATLV01011899 KE524732 KFB36168.1 CH477186 EAT49002.1 UFQS01003074 UFQT01003074 SSX15131.1 SSX34509.1 JXUM01152987 KQ571596 KXJ68155.1 DS231993 EDS30805.1 UFQT01001157 SSX29195.1 UFQS01003844 UFQT01003844 SSX15973.1 SSX35307.1 CCAG010002237 JRES01001582 KNC21736.1 APGK01035007 KB740923 KB632326 ENN78141.1 ERL92426.1 PYGN01000407 PSN46905.1 GAMC01002011 JAC04545.1 NEVH01018377 PNF23593.1 CH933806 EDW13766.1 GEZM01068161 JAV67127.1 KA648856 AFP63485.1 KK118391 KFM72872.1 KK852804 KDR16157.1 CP012526 ALC46510.1 CM000070 EAL29093.2 CH940650 EDW67120.1 CH479185 EDW38405.1 KQ971321 EFA00119.1 OUUW01000007 SPP83421.1 CH916374 EDV91272.1 KK855388 PTY22322.1 AAZX01002214 CVRI01000020 CRK90771.1 CH480815 EDW42636.1 DS235882 EEB20036.1 CM000364 EDX13391.1 PNF23592.1 GBRD01012286 GDHC01017843 GDHC01010739 JAG53538.1 JAQ00786.1 JAQ07890.1 GBHO01010379 JAG33225.1 GGLE01003545 MBY07671.1 GEDC01011430 JAS25868.1 CH954181 EDV49544.1 CM000160 EDW96899.1 CH964232 EDW81221.1 AE014297 BT003495 AAF54616.2 AAO39499.1 CH902617 EDV42978.1 GECZ01007709 JAS62060.1 GDHF01013156 JAI39158.1 GAKP01012483 JAC46469.1 AMQN01001934 KB306824 ELT99474.1 KQ434809 KZC06451.1 KQ761039 OAD58376.1 LJIG01022488 KRT80316.1 CAAE01015019 CAG10690.1 GECU01019152 JAS88554.1 GECL01001121 JAP05003.1 ACPB03006289 GAHY01001664 JAA75846.1 BT057741 ACM09613.1 KQ435767 KOX75284.1 AEMK02000004 CABD030095574 CABD030095575 CABD030095576
KOB79029.1 ODYU01005418 SOQ46235.1 AGBW02012905 OWR44230.1 GAIX01012076 JAA80484.1 KQ459765 KPJ20045.1 KQ459604 KPI92531.1 GDAI01002628 JAI14975.1 GGFJ01007726 MBW56867.1 AXCM01007073 APCN01003107 GGFJ01007875 MBW57016.1 AAAB01008844 EAA06109.3 ADMH02001520 ETN62214.1 AXCN02000227 GAPW01003306 JAC10292.1 ATLV01011899 KE524732 KFB36168.1 CH477186 EAT49002.1 UFQS01003074 UFQT01003074 SSX15131.1 SSX34509.1 JXUM01152987 KQ571596 KXJ68155.1 DS231993 EDS30805.1 UFQT01001157 SSX29195.1 UFQS01003844 UFQT01003844 SSX15973.1 SSX35307.1 CCAG010002237 JRES01001582 KNC21736.1 APGK01035007 KB740923 KB632326 ENN78141.1 ERL92426.1 PYGN01000407 PSN46905.1 GAMC01002011 JAC04545.1 NEVH01018377 PNF23593.1 CH933806 EDW13766.1 GEZM01068161 JAV67127.1 KA648856 AFP63485.1 KK118391 KFM72872.1 KK852804 KDR16157.1 CP012526 ALC46510.1 CM000070 EAL29093.2 CH940650 EDW67120.1 CH479185 EDW38405.1 KQ971321 EFA00119.1 OUUW01000007 SPP83421.1 CH916374 EDV91272.1 KK855388 PTY22322.1 AAZX01002214 CVRI01000020 CRK90771.1 CH480815 EDW42636.1 DS235882 EEB20036.1 CM000364 EDX13391.1 PNF23592.1 GBRD01012286 GDHC01017843 GDHC01010739 JAG53538.1 JAQ00786.1 JAQ07890.1 GBHO01010379 JAG33225.1 GGLE01003545 MBY07671.1 GEDC01011430 JAS25868.1 CH954181 EDV49544.1 CM000160 EDW96899.1 CH964232 EDW81221.1 AE014297 BT003495 AAF54616.2 AAO39499.1 CH902617 EDV42978.1 GECZ01007709 JAS62060.1 GDHF01013156 JAI39158.1 GAKP01012483 JAC46469.1 AMQN01001934 KB306824 ELT99474.1 KQ434809 KZC06451.1 KQ761039 OAD58376.1 LJIG01022488 KRT80316.1 CAAE01015019 CAG10690.1 GECU01019152 JAS88554.1 GECL01001121 JAP05003.1 ACPB03006289 GAHY01001664 JAA75846.1 BT057741 ACM09613.1 KQ435767 KOX75284.1 AEMK02000004 CABD030095574 CABD030095575 CABD030095576
Proteomes
UP000005204
UP000218220
UP000037510
UP000007151
UP000053240
UP000053268
+ More
UP000075885 UP000075882 UP000075920 UP000075883 UP000075840 UP000075903 UP000076407 UP000076408 UP000075880 UP000075902 UP000069272 UP000007062 UP000000673 UP000075886 UP000030765 UP000008820 UP000075900 UP000069940 UP000249989 UP000002320 UP000075884 UP000078200 UP000192223 UP000092444 UP000075881 UP000037069 UP000019118 UP000030742 UP000245037 UP000235965 UP000075901 UP000009192 UP000095301 UP000054359 UP000027135 UP000092553 UP000001819 UP000008792 UP000008744 UP000007266 UP000268350 UP000001070 UP000002358 UP000183832 UP000001292 UP000009046 UP000000304 UP000008711 UP000002282 UP000095300 UP000192221 UP000007798 UP000000803 UP000007801 UP000264800 UP000261580 UP000261520 UP000014760 UP000076502 UP000007303 UP000015103 UP000087266 UP000053105 UP000261640 UP000248480 UP000008227 UP000189704 UP000001519 UP000081671 UP000261620
UP000075885 UP000075882 UP000075920 UP000075883 UP000075840 UP000075903 UP000076407 UP000076408 UP000075880 UP000075902 UP000069272 UP000007062 UP000000673 UP000075886 UP000030765 UP000008820 UP000075900 UP000069940 UP000249989 UP000002320 UP000075884 UP000078200 UP000192223 UP000092444 UP000075881 UP000037069 UP000019118 UP000030742 UP000245037 UP000235965 UP000075901 UP000009192 UP000095301 UP000054359 UP000027135 UP000092553 UP000001819 UP000008792 UP000008744 UP000007266 UP000268350 UP000001070 UP000002358 UP000183832 UP000001292 UP000009046 UP000000304 UP000008711 UP000002282 UP000095300 UP000192221 UP000007798 UP000000803 UP000007801 UP000264800 UP000261580 UP000261520 UP000014760 UP000076502 UP000007303 UP000015103 UP000087266 UP000053105 UP000261640 UP000248480 UP000008227 UP000189704 UP000001519 UP000081671 UP000261620
Interpro
Gene 3D
ProteinModelPortal
H6VTQ5
A0A2A4JZL7
A0A0L7LUG6
A0A2H1VZH0
A0A212ERV4
S4NTV3
+ More
A0A194RRU4 A0A194PGL4 A0A0K8TM17 A0A182PU18 A0A182LAM0 A0A2M4BUY2 A0A182VY83 A0A182MG93 A0A182I8Q6 A0A182V816 A0A182WV32 A0A182XWN1 A0A2M4BVA5 A0A182JCN1 A0A182UAK4 A0A182F7V0 Q7QFV0 W5JHC6 A0A182QCL0 A0A023ELY6 A0A084VDX4 Q0C727 A0A336MZA4 A0A182RDB2 A0A182G8Y7 B0WLZ1 A0A336MG67 A0A182NSR1 A0A1A9VPH1 A0A1W4WP52 A0A336LH30 A0A1B0GCV5 A0A182JY39 A0A0L0BP01 N6UBY2 A0A2P8YRN5 W8CDG0 A0A2J7Q4T4 A0A182SNH9 B4KCZ6 A0A1I8M0Y4 A0A1Y1L7S7 T1PJB2 A0A087U683 A0A067R2E0 A0A0M4EQC9 Q293X1 B4LZI1 B4GLI1 D6WH30 A0A3B0KEF2 B4JUV2 A0A2R7WRY2 K7J968 A0A1J1HS15 B4HIC1 E0W330 B4QU60 A0A2J7Q4T1 A0A0K8SJS3 A0A0A9YUI0 A0A2R5LDQ6 A0A1B6DJR7 B3NZH1 B4PM33 A0A1I8QE65 A0A1W4ULR6 B4NBQ7 Q9VGR0 B3LZ51 A0A1B6GI15 A0A0K8VJW1 A0A3Q3GAV0 A0A034VVK0 A0A3Q4GFT0 A0A3B4AB67 R7U0E8 A0A154P3K1 A0A310SDC4 A0A0T6AYX5 Q4RLP2 H3DJA2 A0A1B6INP3 A0A0V0GAC0 R4FMM3 B9EQ16 A0A0N0BGU1 A0A3Q3LD35 A0A2Y9DT46 A0A287ALB8 A0A1U7UF69 A0A2I2YYS6 A0A1S3EMR0 A0A3Q3VNX4
A0A194RRU4 A0A194PGL4 A0A0K8TM17 A0A182PU18 A0A182LAM0 A0A2M4BUY2 A0A182VY83 A0A182MG93 A0A182I8Q6 A0A182V816 A0A182WV32 A0A182XWN1 A0A2M4BVA5 A0A182JCN1 A0A182UAK4 A0A182F7V0 Q7QFV0 W5JHC6 A0A182QCL0 A0A023ELY6 A0A084VDX4 Q0C727 A0A336MZA4 A0A182RDB2 A0A182G8Y7 B0WLZ1 A0A336MG67 A0A182NSR1 A0A1A9VPH1 A0A1W4WP52 A0A336LH30 A0A1B0GCV5 A0A182JY39 A0A0L0BP01 N6UBY2 A0A2P8YRN5 W8CDG0 A0A2J7Q4T4 A0A182SNH9 B4KCZ6 A0A1I8M0Y4 A0A1Y1L7S7 T1PJB2 A0A087U683 A0A067R2E0 A0A0M4EQC9 Q293X1 B4LZI1 B4GLI1 D6WH30 A0A3B0KEF2 B4JUV2 A0A2R7WRY2 K7J968 A0A1J1HS15 B4HIC1 E0W330 B4QU60 A0A2J7Q4T1 A0A0K8SJS3 A0A0A9YUI0 A0A2R5LDQ6 A0A1B6DJR7 B3NZH1 B4PM33 A0A1I8QE65 A0A1W4ULR6 B4NBQ7 Q9VGR0 B3LZ51 A0A1B6GI15 A0A0K8VJW1 A0A3Q3GAV0 A0A034VVK0 A0A3Q4GFT0 A0A3B4AB67 R7U0E8 A0A154P3K1 A0A310SDC4 A0A0T6AYX5 Q4RLP2 H3DJA2 A0A1B6INP3 A0A0V0GAC0 R4FMM3 B9EQ16 A0A0N0BGU1 A0A3Q3LD35 A0A2Y9DT46 A0A287ALB8 A0A1U7UF69 A0A2I2YYS6 A0A1S3EMR0 A0A3Q3VNX4
PDB
2CTW
E-value=1.25176e-12,
Score=174
Ontologies
GO
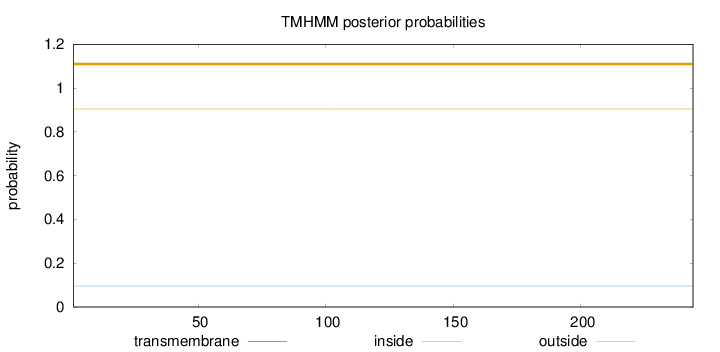
Topology
Length:
244
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0017
Exp number, first 60 AAs:
0.00016
Total prob of N-in:
0.09612
outside
1 - 244
Population Genetic Test Statistics
Pi
38.643714
Theta
49.263493
Tajima's D
-1.109467
CLR
0.657127
CSRT
0.111544422778861
Interpretation
Uncertain
