Gene
KWMTBOMO10960 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008335
Annotation
PREDICTED:_28S_ribosomal_protein_S2?_mitochondrial-like_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 1.031
Sequence
CDS
ATGATTTTGAGTAATACAGGGTGTAGATATACAAAATATGGATTTTCACTTCTGAGATACACATCAACTTCTCCACAAGTCATTCCAGAGAATGTAATTGATGTGCAACAAAATCAGAAAAATCCTCTTGATTACCCAGATTATTTTGGTGTACATAATTTATTTACAGTAAAAGACCTATTTGATGCAAGAGTGCATTATGGTCATAAAATAGGGTCCCTTAATGATAACATGAGATCATACATATATGGTTCAAGACTTGGACACCTCATTTTTGATTTGGATACAACAGCTGAACATTTACGTAAAGCTCTTAATTTCACAGCCCACATTGCATACAGAGGTGGAATCATTTGTTTTTTTAATAGGAATTCACTAAATGCACATATCGTGGAAAAAACAGCAATGGAATGTGGAGAATATGCTCATACAAGATTCTGGAGAGGTGGTATCTTCACAAATGCCAATCAGCAGTTTGGTGCTATAACAAGATTACCTGACTTATGTATATTTCTTAACACCCAGAATAATATTTTGAATCAGCACACAGCTGTCAGAGATAGTGCAAAAATGTTAATACCAACAATTGGAATTGTCGACTCAAATTGTAATCCTAATTTGGTATCATACCCTGTACCTGGCAATGATGATTCCCCAACAGCCATAGAGCTCTACTGTAAGTTATTTAAAAGTGCTATTATTAGAGGAAAGGAAGAAAGACGTAAGTTTCTACAGGAGACAGAGTAA
Protein
MILSNTGCRYTKYGFSLLRYTSTSPQVIPENVIDVQQNQKNPLDYPDYFGVHNLFTVKDLFDARVHYGHKIGSLNDNMRSYIYGSRLGHLIFDLDTTAEHLRKALNFTAHIAYRGGIICFFNRNSLNAHIVEKTAMECGEYAHTRFWRGGIFTNANQQFGAITRLPDLCIFLNTQNNILNQHTAVRDSAKMLIPTIGIVDSNCNPNLVSYPVPGNDDSPTAIELYCKLFKSAIIRGKEERRKFLQETE
Summary
Uniprot
A0A2A4JYU3
A0A2H1W163
A0A0L7LU65
A0A194PGV4
A0A212ERY9
A0A194RV51
+ More
A0A1I8QC64 A0A0L0CDN1 A0A1I8MP94 A0A034VA12 A0A0K8VUV2 W8CBQ4 A0A1B0AGW2 D3TPQ3 A0A1A9YG85 A0A1W4UIX2 A0A3B0K6M5 A0A0M3QT82 A0A1A9VXS9 A0A1A9WMC1 A0A1B0APU0 B3MVJ1 B4NZ19 B4Q337 Q8MSS7 Q29M94 B4G9K0 B4KGQ2 B4N0J9 B4I173 B3N4A7 B4JQB0 B4MDQ0 A0A139WLL3 A0A023EKT4 Q16YD1 A0A0K8TRS0 I4DL61 A0A1S4FJW4 A0A2M4AXZ4 A0A1B0EYW2 A0A182VHR2 A0A182GG99 A0A2C9GQE5 B0X2T8 A0A1S4H7U9 A0A087YU66 W5JUD0 A0A182S839 A0A182TVA5 A0A182J9J0 A0A1Q3F9V5 Q7PZF7 U5ER31 A0A1Y1MNU6 A0A1L8DRC8 K7INN1 A0A182Y9F2 A0A182MTG3 A0A182WDX9 A0A2J7R1Q3 A0A1W4WY49 A0A182JRB1 A0A182XJV1 A0A182L522 A0A232EIX3 A0A182RRZ7 A0A182PKE3 A0A336K5H6 A0A182Q9R8 N6TE21 A0A182N2W7 A0A2P8YET5 A0A3R7M5S7 A0A1B0C9L5 A0A067QZ31 A0A0C9RBN8 A0A1B0FLU6 E9J4W6 A0A158NWW8 F4WVM9 A0A0L7QUF9 A0A195BRU2 A0A1B6L498 A0A2A3ELL6 A0A0P4WVB6 A0A1B6DFW9 A0A151ISA6 R4WE33 A0A195CWX4 A0A088A204 A0A151XK77 A0A026X414 A0A151JYA4 A0A1B6GLH6 A0A182FB46 A0A1B6IXB3 E2BJN7 A0A3R7PWC5
A0A1I8QC64 A0A0L0CDN1 A0A1I8MP94 A0A034VA12 A0A0K8VUV2 W8CBQ4 A0A1B0AGW2 D3TPQ3 A0A1A9YG85 A0A1W4UIX2 A0A3B0K6M5 A0A0M3QT82 A0A1A9VXS9 A0A1A9WMC1 A0A1B0APU0 B3MVJ1 B4NZ19 B4Q337 Q8MSS7 Q29M94 B4G9K0 B4KGQ2 B4N0J9 B4I173 B3N4A7 B4JQB0 B4MDQ0 A0A139WLL3 A0A023EKT4 Q16YD1 A0A0K8TRS0 I4DL61 A0A1S4FJW4 A0A2M4AXZ4 A0A1B0EYW2 A0A182VHR2 A0A182GG99 A0A2C9GQE5 B0X2T8 A0A1S4H7U9 A0A087YU66 W5JUD0 A0A182S839 A0A182TVA5 A0A182J9J0 A0A1Q3F9V5 Q7PZF7 U5ER31 A0A1Y1MNU6 A0A1L8DRC8 K7INN1 A0A182Y9F2 A0A182MTG3 A0A182WDX9 A0A2J7R1Q3 A0A1W4WY49 A0A182JRB1 A0A182XJV1 A0A182L522 A0A232EIX3 A0A182RRZ7 A0A182PKE3 A0A336K5H6 A0A182Q9R8 N6TE21 A0A182N2W7 A0A2P8YET5 A0A3R7M5S7 A0A1B0C9L5 A0A067QZ31 A0A0C9RBN8 A0A1B0FLU6 E9J4W6 A0A158NWW8 F4WVM9 A0A0L7QUF9 A0A195BRU2 A0A1B6L498 A0A2A3ELL6 A0A0P4WVB6 A0A1B6DFW9 A0A151ISA6 R4WE33 A0A195CWX4 A0A088A204 A0A151XK77 A0A026X414 A0A151JYA4 A0A1B6GLH6 A0A182FB46 A0A1B6IXB3 E2BJN7 A0A3R7PWC5
Pubmed
26227816
26354079
22118469
26108605
25315136
25348373
+ More
24495485 20353571 17994087 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 18362917 19820115 24945155 17510324 26369729 22651552 26483478 12364791 20920257 28004739 20075255 25244985 20966253 28648823 23537049 29403074 24845553 21282665 21347285 21719571 23691247 24508170 30249741 20798317
24495485 20353571 17994087 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 18362917 19820115 24945155 17510324 26369729 22651552 26483478 12364791 20920257 28004739 20075255 25244985 20966253 28648823 23537049 29403074 24845553 21282665 21347285 21719571 23691247 24508170 30249741 20798317
EMBL
NWSH01000347
PCG77185.1
ODYU01005418
SOQ46234.1
JTDY01000072
KOB79023.1
+ More
KQ459604 KPI92532.1 AGBW02012905 OWR44229.1 KQ459765 KPJ20046.1 JRES01000536 KNC30356.1 GAKP01020568 GAKP01020565 GAKP01020562 GAKP01020559 GAKP01020556 GAKP01020553 GAKP01020550 GAKP01020547 JAC38393.1 GDHF01009999 JAI42315.1 GAMC01000279 JAC06277.1 EZ423405 ADD19681.1 OUUW01000004 SPP79138.1 CP012523 ALC38429.1 JXJN01001573 JXJN01001574 CH902624 EDV33256.1 CM000157 EDW87683.1 CM000361 CM002910 EDX03753.1 KMY88139.1 AE014134 AY118630 AAF50970.2 AAM49999.1 CH379060 EAL33800.1 CH479180 EDW29030.1 CH933807 EDW13252.1 CH963920 EDW77612.1 CH480820 EDW54280.1 CH954177 EDV57777.1 CH916372 EDV99090.1 CH940661 EDW71311.1 KQ971321 KYB28774.1 GAPW01003818 JAC09780.1 CH477518 EAT39639.1 GDAI01000539 JAI17064.1 AK402029 BAM18651.1 GGFK01012342 MBW45663.1 AJVK01035434 JXUM01061327 JXUM01137733 KQ568620 KQ562145 KXJ68945.1 KXJ76578.1 APCN01002416 DS232298 EDS39412.1 AAAB01008986 ADMH02000120 ETN67751.1 GFDL01010723 JAV24322.1 EAA00165.4 GANO01003954 JAB55917.1 GEZM01031552 JAV84897.1 GFDF01005062 JAV09022.1 AXCM01009764 NEVH01008202 PNF34766.1 NNAY01004149 OXU18295.1 UFQS01000035 UFQT01000035 SSW97953.1 SSX18339.1 AXCN02001090 APGK01033087 KB740860 KB631777 ENN78599.1 ERL86026.1 PYGN01000655 PSN42751.1 QCYY01002111 ROT72797.1 AJWK01002461 AJWK01002462 AJWK01002463 KK852818 KDR15668.1 GBYB01004291 JAG74058.1 CCAG010004762 GL768121 EFZ12121.1 ADTU01028694 GL888391 EGI61747.1 KQ414735 KOC62278.1 KQ976421 KYM89082.1 GEBQ01021497 JAT18480.1 KZ288215 PBC32607.1 GDRN01056185 JAI65860.1 GEDC01012710 JAS24588.1 KQ981090 KYN09566.1 AK418054 BAN21269.1 KQ977171 KYN05145.1 KQ982045 KYQ60737.1 KK107019 QOIP01000007 EZA62751.1 RLU20527.1 KQ981486 KYN41433.1 GECZ01006606 JAS63163.1 GECU01016184 JAS91522.1 GL448604 EFN84095.1 QCYY01001287 ROT79139.1
KQ459604 KPI92532.1 AGBW02012905 OWR44229.1 KQ459765 KPJ20046.1 JRES01000536 KNC30356.1 GAKP01020568 GAKP01020565 GAKP01020562 GAKP01020559 GAKP01020556 GAKP01020553 GAKP01020550 GAKP01020547 JAC38393.1 GDHF01009999 JAI42315.1 GAMC01000279 JAC06277.1 EZ423405 ADD19681.1 OUUW01000004 SPP79138.1 CP012523 ALC38429.1 JXJN01001573 JXJN01001574 CH902624 EDV33256.1 CM000157 EDW87683.1 CM000361 CM002910 EDX03753.1 KMY88139.1 AE014134 AY118630 AAF50970.2 AAM49999.1 CH379060 EAL33800.1 CH479180 EDW29030.1 CH933807 EDW13252.1 CH963920 EDW77612.1 CH480820 EDW54280.1 CH954177 EDV57777.1 CH916372 EDV99090.1 CH940661 EDW71311.1 KQ971321 KYB28774.1 GAPW01003818 JAC09780.1 CH477518 EAT39639.1 GDAI01000539 JAI17064.1 AK402029 BAM18651.1 GGFK01012342 MBW45663.1 AJVK01035434 JXUM01061327 JXUM01137733 KQ568620 KQ562145 KXJ68945.1 KXJ76578.1 APCN01002416 DS232298 EDS39412.1 AAAB01008986 ADMH02000120 ETN67751.1 GFDL01010723 JAV24322.1 EAA00165.4 GANO01003954 JAB55917.1 GEZM01031552 JAV84897.1 GFDF01005062 JAV09022.1 AXCM01009764 NEVH01008202 PNF34766.1 NNAY01004149 OXU18295.1 UFQS01000035 UFQT01000035 SSW97953.1 SSX18339.1 AXCN02001090 APGK01033087 KB740860 KB631777 ENN78599.1 ERL86026.1 PYGN01000655 PSN42751.1 QCYY01002111 ROT72797.1 AJWK01002461 AJWK01002462 AJWK01002463 KK852818 KDR15668.1 GBYB01004291 JAG74058.1 CCAG010004762 GL768121 EFZ12121.1 ADTU01028694 GL888391 EGI61747.1 KQ414735 KOC62278.1 KQ976421 KYM89082.1 GEBQ01021497 JAT18480.1 KZ288215 PBC32607.1 GDRN01056185 JAI65860.1 GEDC01012710 JAS24588.1 KQ981090 KYN09566.1 AK418054 BAN21269.1 KQ977171 KYN05145.1 KQ982045 KYQ60737.1 KK107019 QOIP01000007 EZA62751.1 RLU20527.1 KQ981486 KYN41433.1 GECZ01006606 JAS63163.1 GECU01016184 JAS91522.1 GL448604 EFN84095.1 QCYY01001287 ROT79139.1
Proteomes
UP000218220
UP000037510
UP000053268
UP000007151
UP000053240
UP000095300
+ More
UP000037069 UP000095301 UP000092445 UP000092443 UP000192221 UP000268350 UP000092553 UP000078200 UP000091820 UP000092460 UP000007801 UP000002282 UP000000304 UP000000803 UP000001819 UP000008744 UP000009192 UP000007798 UP000001292 UP000008711 UP000001070 UP000008792 UP000007266 UP000008820 UP000092462 UP000075903 UP000069940 UP000249989 UP000075840 UP000002320 UP000000673 UP000075901 UP000075902 UP000075880 UP000007062 UP000002358 UP000076408 UP000075883 UP000075920 UP000235965 UP000192223 UP000075881 UP000076407 UP000075882 UP000215335 UP000075900 UP000075885 UP000075886 UP000019118 UP000030742 UP000075884 UP000245037 UP000283509 UP000092461 UP000027135 UP000092444 UP000005205 UP000007755 UP000053825 UP000078540 UP000242457 UP000078492 UP000078542 UP000005203 UP000075809 UP000053097 UP000279307 UP000078541 UP000069272 UP000008237
UP000037069 UP000095301 UP000092445 UP000092443 UP000192221 UP000268350 UP000092553 UP000078200 UP000091820 UP000092460 UP000007801 UP000002282 UP000000304 UP000000803 UP000001819 UP000008744 UP000009192 UP000007798 UP000001292 UP000008711 UP000001070 UP000008792 UP000007266 UP000008820 UP000092462 UP000075903 UP000069940 UP000249989 UP000075840 UP000002320 UP000000673 UP000075901 UP000075902 UP000075880 UP000007062 UP000002358 UP000076408 UP000075883 UP000075920 UP000235965 UP000192223 UP000075881 UP000076407 UP000075882 UP000215335 UP000075900 UP000075885 UP000075886 UP000019118 UP000030742 UP000075884 UP000245037 UP000283509 UP000092461 UP000027135 UP000092444 UP000005205 UP000007755 UP000053825 UP000078540 UP000242457 UP000078492 UP000078542 UP000005203 UP000075809 UP000053097 UP000279307 UP000078541 UP000069272 UP000008237
PRIDE
Pfam
Interpro
IPR005706
Ribosomal_S2_bac/mit/plastid
+ More
IPR001865 Ribosomal_S2
IPR023591 Ribosomal_S2_flav_dom_sf
IPR018130 Ribosomal_S2_CS
IPR017986 WD40_repeat_dom
IPR036322 WD40_repeat_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR037626 NUP37
IPR001680 WD40_repeat
IPR010989 SNARE
IPR000727 T_SNARE_dom
IPR001841 Znf_RING
IPR019786 Zinc_finger_PHD-type_CS
IPR019787 Znf_PHD-finger
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR017907 Znf_RING_CS
IPR011011 Znf_FYVE_PHD
IPR040160 Mxt
IPR004088 KH_dom_type_1
IPR016021 MIF4-like_sf
IPR004087 KH_dom
IPR036612 KH_dom_type_1_sf
IPR035979 RBD_domain_sf
IPR000504 RRM_dom
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR001865 Ribosomal_S2
IPR023591 Ribosomal_S2_flav_dom_sf
IPR018130 Ribosomal_S2_CS
IPR017986 WD40_repeat_dom
IPR036322 WD40_repeat_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR037626 NUP37
IPR001680 WD40_repeat
IPR010989 SNARE
IPR000727 T_SNARE_dom
IPR001841 Znf_RING
IPR019786 Zinc_finger_PHD-type_CS
IPR019787 Znf_PHD-finger
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR017907 Znf_RING_CS
IPR011011 Znf_FYVE_PHD
IPR040160 Mxt
IPR004088 KH_dom_type_1
IPR016021 MIF4-like_sf
IPR004087 KH_dom
IPR036612 KH_dom_type_1_sf
IPR035979 RBD_domain_sf
IPR000504 RRM_dom
IPR012677 Nucleotide-bd_a/b_plait_sf
SUPFAM
CDD
ProteinModelPortal
A0A2A4JYU3
A0A2H1W163
A0A0L7LU65
A0A194PGV4
A0A212ERY9
A0A194RV51
+ More
A0A1I8QC64 A0A0L0CDN1 A0A1I8MP94 A0A034VA12 A0A0K8VUV2 W8CBQ4 A0A1B0AGW2 D3TPQ3 A0A1A9YG85 A0A1W4UIX2 A0A3B0K6M5 A0A0M3QT82 A0A1A9VXS9 A0A1A9WMC1 A0A1B0APU0 B3MVJ1 B4NZ19 B4Q337 Q8MSS7 Q29M94 B4G9K0 B4KGQ2 B4N0J9 B4I173 B3N4A7 B4JQB0 B4MDQ0 A0A139WLL3 A0A023EKT4 Q16YD1 A0A0K8TRS0 I4DL61 A0A1S4FJW4 A0A2M4AXZ4 A0A1B0EYW2 A0A182VHR2 A0A182GG99 A0A2C9GQE5 B0X2T8 A0A1S4H7U9 A0A087YU66 W5JUD0 A0A182S839 A0A182TVA5 A0A182J9J0 A0A1Q3F9V5 Q7PZF7 U5ER31 A0A1Y1MNU6 A0A1L8DRC8 K7INN1 A0A182Y9F2 A0A182MTG3 A0A182WDX9 A0A2J7R1Q3 A0A1W4WY49 A0A182JRB1 A0A182XJV1 A0A182L522 A0A232EIX3 A0A182RRZ7 A0A182PKE3 A0A336K5H6 A0A182Q9R8 N6TE21 A0A182N2W7 A0A2P8YET5 A0A3R7M5S7 A0A1B0C9L5 A0A067QZ31 A0A0C9RBN8 A0A1B0FLU6 E9J4W6 A0A158NWW8 F4WVM9 A0A0L7QUF9 A0A195BRU2 A0A1B6L498 A0A2A3ELL6 A0A0P4WVB6 A0A1B6DFW9 A0A151ISA6 R4WE33 A0A195CWX4 A0A088A204 A0A151XK77 A0A026X414 A0A151JYA4 A0A1B6GLH6 A0A182FB46 A0A1B6IXB3 E2BJN7 A0A3R7PWC5
A0A1I8QC64 A0A0L0CDN1 A0A1I8MP94 A0A034VA12 A0A0K8VUV2 W8CBQ4 A0A1B0AGW2 D3TPQ3 A0A1A9YG85 A0A1W4UIX2 A0A3B0K6M5 A0A0M3QT82 A0A1A9VXS9 A0A1A9WMC1 A0A1B0APU0 B3MVJ1 B4NZ19 B4Q337 Q8MSS7 Q29M94 B4G9K0 B4KGQ2 B4N0J9 B4I173 B3N4A7 B4JQB0 B4MDQ0 A0A139WLL3 A0A023EKT4 Q16YD1 A0A0K8TRS0 I4DL61 A0A1S4FJW4 A0A2M4AXZ4 A0A1B0EYW2 A0A182VHR2 A0A182GG99 A0A2C9GQE5 B0X2T8 A0A1S4H7U9 A0A087YU66 W5JUD0 A0A182S839 A0A182TVA5 A0A182J9J0 A0A1Q3F9V5 Q7PZF7 U5ER31 A0A1Y1MNU6 A0A1L8DRC8 K7INN1 A0A182Y9F2 A0A182MTG3 A0A182WDX9 A0A2J7R1Q3 A0A1W4WY49 A0A182JRB1 A0A182XJV1 A0A182L522 A0A232EIX3 A0A182RRZ7 A0A182PKE3 A0A336K5H6 A0A182Q9R8 N6TE21 A0A182N2W7 A0A2P8YET5 A0A3R7M5S7 A0A1B0C9L5 A0A067QZ31 A0A0C9RBN8 A0A1B0FLU6 E9J4W6 A0A158NWW8 F4WVM9 A0A0L7QUF9 A0A195BRU2 A0A1B6L498 A0A2A3ELL6 A0A0P4WVB6 A0A1B6DFW9 A0A151ISA6 R4WE33 A0A195CWX4 A0A088A204 A0A151XK77 A0A026X414 A0A151JYA4 A0A1B6GLH6 A0A182FB46 A0A1B6IXB3 E2BJN7 A0A3R7PWC5
PDB
6NF8
E-value=1.45082e-71,
Score=683
Ontologies
GO
Topology
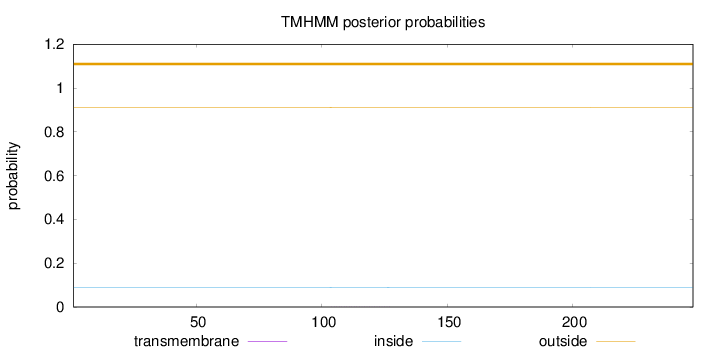
Length:
248
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02263
Exp number, first 60 AAs:
0.00053
Total prob of N-in:
0.08949
outside
1 - 248
Population Genetic Test Statistics
Pi
175.673678
Theta
165.950084
Tajima's D
0.142975
CLR
0.037339
CSRT
0.404129793510324
Interpretation
Uncertain
