Gene
KWMTBOMO10959
Pre Gene Modal
BGIBMGA008504
Annotation
PREDICTED:_leucine-rich_repeat-containing_protein_58-like_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 1.344 Nuclear Reliability : 1.749
Sequence
CDS
ATGATAGATACATTAAGTATCTCTGCTGAAATGAGAAGTATAGTGGATGAAAAAGATTGTGCTGAAAGCTACGAGATAATATTACTGTACAACAACAGAATACAATCACTTCCAAATTTCCTAAACAGGTTTTGCAACTTGAAAATTTTGAATGTGAGCAACAACAGGCTGACTGTTTTACCAGATGTGTTTAAGAATTGTCCCCTCACGACTTTGGTAGCAAAACATAATCAACTCACTAACGAGTCACTTCCCAAATCATTTTATACTGCAAAGAATACTCTTCGTGAACTGAACCTCAGTGGAAATCAGTTAAATTTATTCCCAGAGCAGATATTCGAATTGACACATCTGAAATATCTTTACCTAGGCAGTAATAAAATAGTTAACATACCAAAGGACATTTGGAAATTAAGCGGTTTGCAAATACTATCACTTGGTGGTAACCAAATTATCGAAGTGCCCGAGAACGTAGGTCACTTAACTTCACTACAAGCGTTGGTGCTCAGTGATAATTTGATCGAACAATTGCCAGCAAGCATCACAAAACTGAAGAATTTAAGATCTTTATTATTGCACAAGAATCGATTGAAAACTTTGCCTACACAAATTATTAAACTAAAATGCTTGACAGAGCTCAGTCTTCGGGACAATCCCCTTGTAGTAAGATTCGTTAGAGACATGACTCTGCAGCCACCATCACTCTTAGAGCTGGCTGGTCGCACTGTGAAACTTCACAACATTCCAATTGAAACTGGAGGTATTCCTAAGACATTGATCAATTATCTGCAAGCAGCCCAATGTTGTGTCAATCCAAAATGCAAAGGTGTCTTCTTTGACAACCGCGTAGAACACATCAAATTTGTAGATTTTTGTGGAAAGTACAGGATTCCATTACTACAATATTTATGCAGTTCAAAATGTATTACTGGAAGTTGGGAAAGCCGTGAGGTAGACAGTAATCCTCACCCACATATGATGCGAAAGGTTCTTCTGGGTTAA
Protein
MIDTLSISAEMRSIVDEKDCAESYEIILLYNNRIQSLPNFLNRFCNLKILNVSNNRLTVLPDVFKNCPLTTLVAKHNQLTNESLPKSFYTAKNTLRELNLSGNQLNLFPEQIFELTHLKYLYLGSNKIVNIPKDIWKLSGLQILSLGGNQIIEVPENVGHLTSLQALVLSDNLIEQLPASITKLKNLRSLLLHKNRLKTLPTQIIKLKCLTELSLRDNPLVVRFVRDMTLQPPSLLELAGRTVKLHNIPIETGGIPKTLINYLQAAQCCVNPKCKGVFFDNRVEHIKFVDFCGKYRIPLLQYLCSSKCITGSWESREVDSNPHPHMMRKVLLG
Summary
Uniprot
H9JG57
A0A2A4JZI7
A0A2H1VZL3
A0A194PHK7
A0A3S2TB08
A0A194RRE8
+ More
A0A212ERV7 A0A182J465 A0A182V8P5 F5HLP7 A0A182XFB1 A0A182MQU9 A0A182Y8V9 A0A182RT39 A0A182TBA7 A0A182PTB0 A0A0L0BYP7 A0A2J7R569 A0A182JTY3 A0A182KZM5 A0A182HYK4 A0A1I8PYS4 A0A182UIC7 A0A0K8W0P6 A0A034WHR9 A0A0C9RTW8 A0A0A1WYI1 A0A182W3C8 A0A2M4BSA4 A0A1L8EID8 A0A182F9H7 A0A1L8EHZ7 A0A2M4ACH6 A0A023EQH5 T1DH18 A0A2M4BSN3 A0A2M3YYG7 W5JJ38 A0A1Q3FD35 A0A1Q3FBH1 D3TPL0 T1PDI2 B4NPA7 B3NW23 B0WH12 B4PXX5 A0A2M4A1L0 A0A2M4A1E6 A0A182G6T3 A0A0L7QNV0 A0A088AP30 A0A2A3ERZ4 B3MY09 W8AYH5 A0A2M4AJ77 A0A0N0BDK6 A0A023EQD6 B4L3E1 A0A1A9W3Z5 Q17AP2 B4R7K5 A0A182Q528 A0A3B0KV71 A0A195EJN5 Q9W2U2 Q29H41 A0A195CA71 A0A139WJ18 A0A0M5J1I7 B4H0D1 B4MES5 A0A1L8E9I2 A0A154PNX7 E2C156 A0A1B6DR41 E2A7W2 A0A1W4V7A6 A0A195BGP4 J3JTC7 A0A1B0GFQ0 A0A1B0AC22 A0A151WXN1 B4JKQ0 A0A1A9ULD6 A0A1J1IW72 A0A195F1L7 A0A3L8DS70 F4X3W0 A0A026X1H7 A0A067RNP1 A0A1S3CWN8 T1I634 A0A0P4VR27 E9I8X1 A0A336KQ08 A0A224XRD2 E0VLD6 A0A1J1IU91 A0A069DYR8 A0A1Y1KD58
A0A212ERV7 A0A182J465 A0A182V8P5 F5HLP7 A0A182XFB1 A0A182MQU9 A0A182Y8V9 A0A182RT39 A0A182TBA7 A0A182PTB0 A0A0L0BYP7 A0A2J7R569 A0A182JTY3 A0A182KZM5 A0A182HYK4 A0A1I8PYS4 A0A182UIC7 A0A0K8W0P6 A0A034WHR9 A0A0C9RTW8 A0A0A1WYI1 A0A182W3C8 A0A2M4BSA4 A0A1L8EID8 A0A182F9H7 A0A1L8EHZ7 A0A2M4ACH6 A0A023EQH5 T1DH18 A0A2M4BSN3 A0A2M3YYG7 W5JJ38 A0A1Q3FD35 A0A1Q3FBH1 D3TPL0 T1PDI2 B4NPA7 B3NW23 B0WH12 B4PXX5 A0A2M4A1L0 A0A2M4A1E6 A0A182G6T3 A0A0L7QNV0 A0A088AP30 A0A2A3ERZ4 B3MY09 W8AYH5 A0A2M4AJ77 A0A0N0BDK6 A0A023EQD6 B4L3E1 A0A1A9W3Z5 Q17AP2 B4R7K5 A0A182Q528 A0A3B0KV71 A0A195EJN5 Q9W2U2 Q29H41 A0A195CA71 A0A139WJ18 A0A0M5J1I7 B4H0D1 B4MES5 A0A1L8E9I2 A0A154PNX7 E2C156 A0A1B6DR41 E2A7W2 A0A1W4V7A6 A0A195BGP4 J3JTC7 A0A1B0GFQ0 A0A1B0AC22 A0A151WXN1 B4JKQ0 A0A1A9ULD6 A0A1J1IW72 A0A195F1L7 A0A3L8DS70 F4X3W0 A0A026X1H7 A0A067RNP1 A0A1S3CWN8 T1I634 A0A0P4VR27 E9I8X1 A0A336KQ08 A0A224XRD2 E0VLD6 A0A1J1IU91 A0A069DYR8 A0A1Y1KD58
Pubmed
19121390
26354079
22118469
12364791
14747013
17210077
+ More
25244985 26108605 20966253 25348373 25830018 24945155 20920257 23761445 20353571 25315136 17994087 17550304 26483478 24495485 17510324 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 18362917 19820115 20798317 22516182 23537049 30249741 21719571 24508170 24845553 27129103 21282665 20566863 26334808 28004739
25244985 26108605 20966253 25348373 25830018 24945155 20920257 23761445 20353571 25315136 17994087 17550304 26483478 24495485 17510324 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 18362917 19820115 20798317 22516182 23537049 30249741 21719571 24508170 24845553 27129103 21282665 20566863 26334808 28004739
EMBL
BABH01002039
NWSH01000347
PCG77186.1
ODYU01005418
SOQ46233.1
KQ459604
+ More
KPI92533.1 RSAL01001779 RVE40697.1 KQ459765 KPJ20047.1 AGBW02012905 OWR44228.1 AAAB01008898 EAA44590.4 EGK97180.1 EGK97181.1 AXCM01000209 JRES01001143 KNC25167.1 NEVH01007393 PNF35984.1 APCN01005389 GDHF01007643 JAI44671.1 GAKP01005065 GAKP01005064 JAC53887.1 GBYB01011026 JAG80793.1 GBXI01010829 JAD03463.1 GGFJ01006789 MBW55930.1 GFDG01000446 JAV18353.1 GFDG01000447 JAV18352.1 GGFK01005148 MBW38469.1 GAPW01002245 JAC11353.1 GAMD01002546 JAA99044.1 GGFJ01006790 MBW55931.1 GGFM01000571 MBW21322.1 ADMH02001021 ETN64387.1 GFDL01009612 JAV25433.1 GFDL01010183 JAV24862.1 EZ423362 ADD19638.1 KA646190 AFP60819.1 CH964291 EDW86347.1 CH954180 EDV46156.1 DS231931 EDS27337.1 CM000162 EDX02947.1 GGFK01001314 MBW34635.1 GGFK01001315 MBW34636.1 JXUM01007509 JXUM01007510 KQ560241 KXJ83636.1 KQ414849 KOC60244.1 KZ288193 PBC33986.1 CH902630 EDV38624.1 GAMC01015363 GAMC01015362 GAMC01015361 JAB91192.1 GGFK01007513 MBW40834.1 KQ435859 KOX70562.1 GAPW01002819 JAC10779.1 CH933810 EDW07069.1 CH477331 EAT43344.1 CM000366 EDX17564.1 AXCN02000234 OUUW01000019 SPP89251.1 KQ978782 KYN28465.1 AE014298 AY113577 AAF46596.2 AAM29582.1 CH379064 EAL31918.1 KQ978081 KYM97091.1 KQ971338 KYB27980.1 CP012528 ALC49956.1 CH479200 EDW29726.1 CH940664 EDW63050.1 GFDG01003412 JAV15387.1 KQ435007 KZC13563.1 GL451853 EFN78377.1 GEDC01009140 JAS28158.1 GL437432 EFN70476.1 KQ976490 KYM83391.1 APGK01010787 BT126483 KB738301 KB632179 AEE61447.1 ENN82675.1 ERL89609.1 CCAG010023033 KQ982663 KYQ52608.1 CH916370 EDW00153.1 CVRI01000059 CRL03828.1 KQ981856 KYN34470.1 QOIP01000005 RLU23166.1 GL888624 EGI58907.1 KK107063 EZA61244.1 KK852559 KDR21359.1 ACPB03020305 GDKW01001116 JAI55479.1 GL761662 EFZ22974.1 UFQS01000789 UFQT01000789 SSX06894.1 SSX27238.1 GFTR01005426 JAW11000.1 DS235271 EEB14192.1 CRL03827.1 GBGD01001915 JAC86974.1 GEZM01086441 GEZM01086439 GEZM01086436 JAV59443.1
KPI92533.1 RSAL01001779 RVE40697.1 KQ459765 KPJ20047.1 AGBW02012905 OWR44228.1 AAAB01008898 EAA44590.4 EGK97180.1 EGK97181.1 AXCM01000209 JRES01001143 KNC25167.1 NEVH01007393 PNF35984.1 APCN01005389 GDHF01007643 JAI44671.1 GAKP01005065 GAKP01005064 JAC53887.1 GBYB01011026 JAG80793.1 GBXI01010829 JAD03463.1 GGFJ01006789 MBW55930.1 GFDG01000446 JAV18353.1 GFDG01000447 JAV18352.1 GGFK01005148 MBW38469.1 GAPW01002245 JAC11353.1 GAMD01002546 JAA99044.1 GGFJ01006790 MBW55931.1 GGFM01000571 MBW21322.1 ADMH02001021 ETN64387.1 GFDL01009612 JAV25433.1 GFDL01010183 JAV24862.1 EZ423362 ADD19638.1 KA646190 AFP60819.1 CH964291 EDW86347.1 CH954180 EDV46156.1 DS231931 EDS27337.1 CM000162 EDX02947.1 GGFK01001314 MBW34635.1 GGFK01001315 MBW34636.1 JXUM01007509 JXUM01007510 KQ560241 KXJ83636.1 KQ414849 KOC60244.1 KZ288193 PBC33986.1 CH902630 EDV38624.1 GAMC01015363 GAMC01015362 GAMC01015361 JAB91192.1 GGFK01007513 MBW40834.1 KQ435859 KOX70562.1 GAPW01002819 JAC10779.1 CH933810 EDW07069.1 CH477331 EAT43344.1 CM000366 EDX17564.1 AXCN02000234 OUUW01000019 SPP89251.1 KQ978782 KYN28465.1 AE014298 AY113577 AAF46596.2 AAM29582.1 CH379064 EAL31918.1 KQ978081 KYM97091.1 KQ971338 KYB27980.1 CP012528 ALC49956.1 CH479200 EDW29726.1 CH940664 EDW63050.1 GFDG01003412 JAV15387.1 KQ435007 KZC13563.1 GL451853 EFN78377.1 GEDC01009140 JAS28158.1 GL437432 EFN70476.1 KQ976490 KYM83391.1 APGK01010787 BT126483 KB738301 KB632179 AEE61447.1 ENN82675.1 ERL89609.1 CCAG010023033 KQ982663 KYQ52608.1 CH916370 EDW00153.1 CVRI01000059 CRL03828.1 KQ981856 KYN34470.1 QOIP01000005 RLU23166.1 GL888624 EGI58907.1 KK107063 EZA61244.1 KK852559 KDR21359.1 ACPB03020305 GDKW01001116 JAI55479.1 GL761662 EFZ22974.1 UFQS01000789 UFQT01000789 SSX06894.1 SSX27238.1 GFTR01005426 JAW11000.1 DS235271 EEB14192.1 CRL03827.1 GBGD01001915 JAC86974.1 GEZM01086441 GEZM01086439 GEZM01086436 JAV59443.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000283053
UP000053240
UP000007151
+ More
UP000075880 UP000075903 UP000007062 UP000076407 UP000075883 UP000076408 UP000075900 UP000075901 UP000075885 UP000037069 UP000235965 UP000075881 UP000075882 UP000075840 UP000095300 UP000075902 UP000075920 UP000069272 UP000000673 UP000095301 UP000007798 UP000008711 UP000002320 UP000002282 UP000069940 UP000249989 UP000053825 UP000005203 UP000242457 UP000007801 UP000053105 UP000009192 UP000091820 UP000008820 UP000000304 UP000075886 UP000268350 UP000078492 UP000000803 UP000001819 UP000078542 UP000007266 UP000092553 UP000008744 UP000008792 UP000076502 UP000008237 UP000000311 UP000192221 UP000078540 UP000019118 UP000030742 UP000092444 UP000092445 UP000075809 UP000001070 UP000078200 UP000183832 UP000078541 UP000279307 UP000007755 UP000053097 UP000027135 UP000079169 UP000015103 UP000009046
UP000075880 UP000075903 UP000007062 UP000076407 UP000075883 UP000076408 UP000075900 UP000075901 UP000075885 UP000037069 UP000235965 UP000075881 UP000075882 UP000075840 UP000095300 UP000075902 UP000075920 UP000069272 UP000000673 UP000095301 UP000007798 UP000008711 UP000002320 UP000002282 UP000069940 UP000249989 UP000053825 UP000005203 UP000242457 UP000007801 UP000053105 UP000009192 UP000091820 UP000008820 UP000000304 UP000075886 UP000268350 UP000078492 UP000000803 UP000001819 UP000078542 UP000007266 UP000092553 UP000008744 UP000008792 UP000076502 UP000008237 UP000000311 UP000192221 UP000078540 UP000019118 UP000030742 UP000092444 UP000092445 UP000075809 UP000001070 UP000078200 UP000183832 UP000078541 UP000279307 UP000007755 UP000053097 UP000027135 UP000079169 UP000015103 UP000009046
Interpro
SUPFAM
SSF53098
SSF53098
Gene 3D
ProteinModelPortal
H9JG57
A0A2A4JZI7
A0A2H1VZL3
A0A194PHK7
A0A3S2TB08
A0A194RRE8
+ More
A0A212ERV7 A0A182J465 A0A182V8P5 F5HLP7 A0A182XFB1 A0A182MQU9 A0A182Y8V9 A0A182RT39 A0A182TBA7 A0A182PTB0 A0A0L0BYP7 A0A2J7R569 A0A182JTY3 A0A182KZM5 A0A182HYK4 A0A1I8PYS4 A0A182UIC7 A0A0K8W0P6 A0A034WHR9 A0A0C9RTW8 A0A0A1WYI1 A0A182W3C8 A0A2M4BSA4 A0A1L8EID8 A0A182F9H7 A0A1L8EHZ7 A0A2M4ACH6 A0A023EQH5 T1DH18 A0A2M4BSN3 A0A2M3YYG7 W5JJ38 A0A1Q3FD35 A0A1Q3FBH1 D3TPL0 T1PDI2 B4NPA7 B3NW23 B0WH12 B4PXX5 A0A2M4A1L0 A0A2M4A1E6 A0A182G6T3 A0A0L7QNV0 A0A088AP30 A0A2A3ERZ4 B3MY09 W8AYH5 A0A2M4AJ77 A0A0N0BDK6 A0A023EQD6 B4L3E1 A0A1A9W3Z5 Q17AP2 B4R7K5 A0A182Q528 A0A3B0KV71 A0A195EJN5 Q9W2U2 Q29H41 A0A195CA71 A0A139WJ18 A0A0M5J1I7 B4H0D1 B4MES5 A0A1L8E9I2 A0A154PNX7 E2C156 A0A1B6DR41 E2A7W2 A0A1W4V7A6 A0A195BGP4 J3JTC7 A0A1B0GFQ0 A0A1B0AC22 A0A151WXN1 B4JKQ0 A0A1A9ULD6 A0A1J1IW72 A0A195F1L7 A0A3L8DS70 F4X3W0 A0A026X1H7 A0A067RNP1 A0A1S3CWN8 T1I634 A0A0P4VR27 E9I8X1 A0A336KQ08 A0A224XRD2 E0VLD6 A0A1J1IU91 A0A069DYR8 A0A1Y1KD58
A0A212ERV7 A0A182J465 A0A182V8P5 F5HLP7 A0A182XFB1 A0A182MQU9 A0A182Y8V9 A0A182RT39 A0A182TBA7 A0A182PTB0 A0A0L0BYP7 A0A2J7R569 A0A182JTY3 A0A182KZM5 A0A182HYK4 A0A1I8PYS4 A0A182UIC7 A0A0K8W0P6 A0A034WHR9 A0A0C9RTW8 A0A0A1WYI1 A0A182W3C8 A0A2M4BSA4 A0A1L8EID8 A0A182F9H7 A0A1L8EHZ7 A0A2M4ACH6 A0A023EQH5 T1DH18 A0A2M4BSN3 A0A2M3YYG7 W5JJ38 A0A1Q3FD35 A0A1Q3FBH1 D3TPL0 T1PDI2 B4NPA7 B3NW23 B0WH12 B4PXX5 A0A2M4A1L0 A0A2M4A1E6 A0A182G6T3 A0A0L7QNV0 A0A088AP30 A0A2A3ERZ4 B3MY09 W8AYH5 A0A2M4AJ77 A0A0N0BDK6 A0A023EQD6 B4L3E1 A0A1A9W3Z5 Q17AP2 B4R7K5 A0A182Q528 A0A3B0KV71 A0A195EJN5 Q9W2U2 Q29H41 A0A195CA71 A0A139WJ18 A0A0M5J1I7 B4H0D1 B4MES5 A0A1L8E9I2 A0A154PNX7 E2C156 A0A1B6DR41 E2A7W2 A0A1W4V7A6 A0A195BGP4 J3JTC7 A0A1B0GFQ0 A0A1B0AC22 A0A151WXN1 B4JKQ0 A0A1A9ULD6 A0A1J1IW72 A0A195F1L7 A0A3L8DS70 F4X3W0 A0A026X1H7 A0A067RNP1 A0A1S3CWN8 T1I634 A0A0P4VR27 E9I8X1 A0A336KQ08 A0A224XRD2 E0VLD6 A0A1J1IU91 A0A069DYR8 A0A1Y1KD58
PDB
4U06
E-value=3.22471e-25,
Score=285
Ontologies
GO
Topology
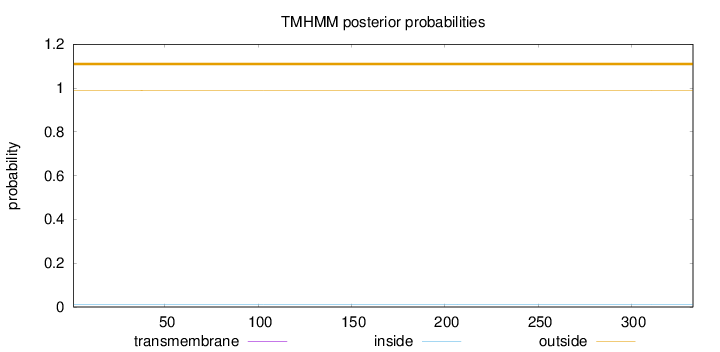
Length:
333
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000620000000000001
Exp number, first 60 AAs:
0.00027
Total prob of N-in:
0.01179
outside
1 - 333
Population Genetic Test Statistics
Pi
233.939702
Theta
198.388353
Tajima's D
0.100606
CLR
0.266113
CSRT
0.396530173491325
Interpretation
Uncertain
