Gene
KWMTBOMO10958 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008336
Annotation
PREDICTED:_glycogen_synthase_kinase-3_beta_isoform_X3_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.218 Mitochondrial Reliability : 1.204 Nuclear Reliability : 1.574
Sequence
CDS
ATGAGCGGACGCCCTAGGACCACGTCATTCGCCGAGGGCAGCAAATCCGTTCGAAAGACTTTCGACAATCAACCTCCAAAACCACCTCTCGGAGGCGTAAAAATTAGCAGCAAGGACGGCTCAAAGGTGACGACTGTGGTAGCGACGCCGGGGCAGGGCCCTGACCGGCCTCAGGAAGTGAGCTATGCTGACATGAAATTGATCGGCAACGGCAGCTTCGGCGTCGTCTATCAAGCCAAACTATGTGACACCGGCGAGCTCATTGCCATCAAGAAGGTGCTCCAGGACAAGAGGTTTAAGAACCGAGAACTTCAAATCATGCGACGGCTGGAGCATTGCAACATTGTGAAACTGAAGTATTTCTTTTATTCCAGTGGCGAAAAGAAAGACGAAGTGTACCTGAATTTGGTGCTGGAGTACATACCCGAGACCGTGTACAAAGTGGCGCGTCATTACTCCAAAGACGAACAAACCATACCCATTAGTTTTATAAAGCTGTACATGTACCAGCTGTTCAGAAGTCTGGCGTACATTCATTCTTTGGGCATCTGTCACCGAGATATCAAGCCGCAGAACCTATTGCTTGACCCCAAGACTGGAGTGTTGAAGCTGTGCGACTTTGGCTCGGCGAAGCACCTCGTTCGCGGAGAACCAAACGTTTCCTACATATGTTCGCGTTATTACCGCGCTCCGGAGCTCATCTTTGGCGCCACCGACTACACCACGAAAATTGACGTGTGGAGCGCGGGATGCGTTGTCGCTGAGCTGTTGTTGGGTCAACCGATATTCCCAGGAGATTCCGGGGTTGACCAATTGGTTGAAATTATCAAGGTTTTAGGGACACCGACAAGAGAACAAATCCGAGAGATGAATCCAAATTATACAGAATTTAAGTTTCCGCAAATCAAGAGTCACCCTTGGGCGAAGGTGTTCCGCGCGTGCACGCCCCCGGACGCCATCTCGCTGGTGTCGCGGCTGCTGGAGTACACGCCGGGCGCGCGCCTGTCGCCGCTGCAGGCCTGCGTGCACAGCTTCTTCGACGAGCTGCGCGAGCCCACCGCCCGCCTGCCCAACGGCCGCCCGCTGCCGCCGCTCTTCAACTTCACCGAGTACGAGCTCGGCATCCAGCCCTCGCTCAACGAGTTCCTGAAGCCGCGCAGCGCCGCGCAGTCCGAGCCGGCCGCCGCCTCCTCTGCGCCCGACCACGACGCGCCAGGCGAGGCGGCATCGGGCGGCGCGACGGCCGACGCTTCCTAG
Protein
MSGRPRTTSFAEGSKSVRKTFDNQPPKPPLGGVKISSKDGSKVTTVVATPGQGPDRPQEVSYADMKLIGNGSFGVVYQAKLCDTGELIAIKKVLQDKRFKNRELQIMRRLEHCNIVKLKYFFYSSGEKKDEVYLNLVLEYIPETVYKVARHYSKDEQTIPISFIKLYMYQLFRSLAYIHSLGICHRDIKPQNLLLDPKTGVLKLCDFGSAKHLVRGEPNVSYICSRYYRAPELIFGATDYTTKIDVWSAGCVVAELLLGQPIFPGDSGVDQLVEIIKVLGTPTREQIREMNPNYTEFKFPQIKSHPWAKVFRACTPPDAISLVSRLLEYTPGARLSPLQACVHSFFDELREPTARLPNGRPLPPLFNFTEYELGIQPSLNEFLKPRSAAQSEPAAASSAPDHDAPGEAASGGATADAS
Summary
Similarity
Belongs to the protein kinase superfamily.
Uniprot
A0A3S2NX33
A0A2A4JYU5
A0A0N7B1F3
S4P264
B0LBZ5
A0A2H1VZH2
+ More
A0A1B6C0U9 A0A1B6F6J0 T1H8J1 A0A194RV56 A0A0V0G765 A0A023F639 A0A1Y1KDH6 A0A0K8S854 A0A1L5YQK4 A0A0M4N2C8 A0A1B6GHH2 G3M3K4 A0A1Y1KKT3 A0A336KSE0 A0A1L8DIV1 A0A1L8DKQ2 A0A0C9R1V4 A0A1B6D5Y6 A0A2R5L9D4 U5EWA5 A0A2M4A5A8 A0A1S6LQ19 Q1HRP9 A0A1Q3G2A6 A0A1Q3G280 T1DE55 A0A1B6FQA7 V5HHR9 A0A0C9QU91 A0A286RXX4 A0A2I6BQ08 F5HLN9 V9IA57 A0A1B6E4X9 A0A087UXA8 A0A1B6EFD5 A0A0A9X6R6 A0A2J7R578 A9P773 A0A1E1X322 A0A023GMQ4 A0A1E1XS44 A0A023FX91 A0A023FIX7 A0A2L2YB23 T1IIH7 L7M8D6 A0A224YNQ5 A0A131Z5I0 A0A0P6C028 A0A067RL75 A0A0K8TM77 A0A069DSV4 A0A131XBN5 A0A0P5KKG9 A0A1B6FDU2 V9ICE0 A0A0P5ZK17 A0A034VLD1 A0A0P5I123 A0A0P5TXZ5 A0A2H8U153 A0A2S2PQD5 B5THN8 A0A0P4YJ16 E9H6Q6 B4JNN7 A0A0A9XG80 A0A1B6MI16 A0A034VMD5 A0A2P2I2X3 C3Y0G1 A0A1I9P1M5 W8C918 A0A0K8S849 A0A0K8VES1 A0A087UB97 A0A1I9WLM5 A0A1Y1KGF5 A0A2M4CZ54 B4M217 T1PFF3 A0A218VDR1 A0A1V4JB40 A0A3Q2TVV5 A0A2I0MLE2 H0ZT29 A0A0Q9WLE3 A0A0Q9XGA1 W5MJG8 A0A0P6IRZ3 A0A026WZI6 A0A2U9C9M7 A0A151MD28
A0A1B6C0U9 A0A1B6F6J0 T1H8J1 A0A194RV56 A0A0V0G765 A0A023F639 A0A1Y1KDH6 A0A0K8S854 A0A1L5YQK4 A0A0M4N2C8 A0A1B6GHH2 G3M3K4 A0A1Y1KKT3 A0A336KSE0 A0A1L8DIV1 A0A1L8DKQ2 A0A0C9R1V4 A0A1B6D5Y6 A0A2R5L9D4 U5EWA5 A0A2M4A5A8 A0A1S6LQ19 Q1HRP9 A0A1Q3G2A6 A0A1Q3G280 T1DE55 A0A1B6FQA7 V5HHR9 A0A0C9QU91 A0A286RXX4 A0A2I6BQ08 F5HLN9 V9IA57 A0A1B6E4X9 A0A087UXA8 A0A1B6EFD5 A0A0A9X6R6 A0A2J7R578 A9P773 A0A1E1X322 A0A023GMQ4 A0A1E1XS44 A0A023FX91 A0A023FIX7 A0A2L2YB23 T1IIH7 L7M8D6 A0A224YNQ5 A0A131Z5I0 A0A0P6C028 A0A067RL75 A0A0K8TM77 A0A069DSV4 A0A131XBN5 A0A0P5KKG9 A0A1B6FDU2 V9ICE0 A0A0P5ZK17 A0A034VLD1 A0A0P5I123 A0A0P5TXZ5 A0A2H8U153 A0A2S2PQD5 B5THN8 A0A0P4YJ16 E9H6Q6 B4JNN7 A0A0A9XG80 A0A1B6MI16 A0A034VMD5 A0A2P2I2X3 C3Y0G1 A0A1I9P1M5 W8C918 A0A0K8S849 A0A0K8VES1 A0A087UB97 A0A1I9WLM5 A0A1Y1KGF5 A0A2M4CZ54 B4M217 T1PFF3 A0A218VDR1 A0A1V4JB40 A0A3Q2TVV5 A0A2I0MLE2 H0ZT29 A0A0Q9WLE3 A0A0Q9XGA1 W5MJG8 A0A0P6IRZ3 A0A026WZI6 A0A2U9C9M7 A0A151MD28
Pubmed
23622113
18184036
18183285
26354079
25474469
28004739
+ More
26823975 18362917 19820115 17204158 24330624 25765539 29255161 12364791 14747013 17210077 25401762 19285806 28503490 29209593 26561354 25576852 28797301 26830274 24845553 26369729 26334808 28049606 25348373 21292972 17994087 18563158 24495485 27538518 15592404 23371554 20360741 24508170 22293439
26823975 18362917 19820115 17204158 24330624 25765539 29255161 12364791 14747013 17210077 25401762 19285806 28503490 29209593 26561354 25576852 28797301 26830274 24845553 26369729 26334808 28049606 25348373 21292972 17994087 18563158 24495485 27538518 15592404 23371554 20360741 24508170 22293439
EMBL
RSAL01000119
RVE46742.1
NWSH01000347
PCG77187.1
KM233161
AKC42322.1
+ More
GAIX01012175 JAA80385.1 EF554581 ABU49716.1 ODYU01005418 SOQ46230.1 GEDC01030304 GEDC01010866 JAS06994.1 JAS26432.1 GECZ01023949 JAS45820.1 ACPB03010082 KQ459765 KPJ20051.1 GECL01002323 JAP03801.1 GBBI01002044 JAC16668.1 GEZM01086439 GEZM01086438 JAV59444.1 GBRD01016483 GBRD01016482 GDHC01012482 JAG49344.1 JAQ06147.1 KX602320 APP94029.1 KR075840 ALE20559.1 GECZ01007892 JAS61877.1 JN602375 KQ971338 AEO44887.1 KYB27979.1 GEZM01086441 GEZM01086437 JAV59437.1 UFQS01000789 UFQT01000789 SSX06891.1 SSX27235.1 GFDF01007701 JAV06383.1 GFDF01007164 JAV06920.1 GBYB01000831 JAG70598.1 GEDC01016201 JAS21097.1 GGLE01001962 MBY06088.1 GANO01003038 JAB56833.1 GGFK01002589 MBW35910.1 KU641425 MG680178 AQT40602.1 AWG41959.1 DQ440045 ABF18078.1 GFDL01001112 JAV33933.1 GFDL01001118 JAV33927.1 GALA01001182 JAA93670.1 GECZ01017425 JAS52344.1 GANP01011440 JAB73028.1 GBYB01007299 JAG77066.1 KY974307 ASW35107.1 KY923001 AUI80372.1 AAAB01008898 EGK97172.1 JR037830 AEY57993.1 GEDC01004317 JAS32981.1 KK122137 KFM81997.1 GEDC01019129 GEDC01000724 JAS18169.1 JAS36574.1 GBHO01029101 GBHO01025784 GBHO01017337 GBRD01016489 GBRD01016486 JAG14503.1 JAG17820.1 JAG26267.1 JAG49337.1 NEVH01007393 PNF35983.1 EF142066 ABO61882.1 GFAC01005508 JAT93680.1 GBBM01001268 JAC34150.1 GFAA01001287 JAU02148.1 GBBL01001221 JAC26099.1 GBBK01002606 JAC21876.1 IAAA01011464 LAA05187.1 AFFK01014267 GACK01004729 JAA60305.1 GFPF01008092 MAA19238.1 GEDV01001980 JAP86577.1 GDIP01007833 JAM95882.1 KK852559 KDR21360.1 GDAI01002144 JAI15459.1 GBGD01001716 JAC87173.1 GEFH01004028 JAP64553.1 GDIQ01184278 JAK67447.1 GECZ01021409 JAS48360.1 JR037831 AEY57994.1 GDIP01043960 JAM59755.1 GAKP01015698 JAC43254.1 GDIQ01225908 GDIP01112240 JAK25817.1 JAL91474.1 GDIP01120453 JAL83261.1 GFXV01007333 MBW19138.1 GGMR01018437 MBY31056.1 EU939764 ACH73246.1 GDIP01226788 JAI96613.1 GL732598 EFX72595.1 CH916371 EDV92330.1 GBHO01035195 GBHO01025781 GBHO01018235 GBRD01016485 GBRD01007877 JAG08409.1 JAG17823.1 JAG25369.1 JAG49341.1 GEBQ01004458 JAT35519.1 GAKP01015695 JAC43257.1 IACF01002588 LAB68236.1 GG666478 EEN66234.1 KT336322 ANZ22981.1 GAMC01007026 GAMC01007025 JAB99529.1 GBRD01016487 GBRD01007876 GDHC01015893 JAG49339.1 JAQ02736.1 GDHF01015274 JAI37040.1 KK119083 KFM74636.1 KU932409 APA34045.1 GEZM01086440 GEZM01086436 JAV59451.1 GGFL01006367 MBW70545.1 CH940651 EDW65721.1 KA646638 AFP61267.1 MUZQ01000007 OWK63988.1 LSYS01008075 OPJ69398.1 AADN05000813 AKCR02000007 PKK30500.1 ABQF01034770 ABQF01034771 ABQF01034772 ABQF01034773 KRF82353.1 CH933815 KRG07531.1 AHAT01020299 AHAT01020300 GDIQ01006531 JAN88206.1 KK107063 EZA61243.1 CP026256 AWP13254.1 AKHW03006231 KYO22436.1
GAIX01012175 JAA80385.1 EF554581 ABU49716.1 ODYU01005418 SOQ46230.1 GEDC01030304 GEDC01010866 JAS06994.1 JAS26432.1 GECZ01023949 JAS45820.1 ACPB03010082 KQ459765 KPJ20051.1 GECL01002323 JAP03801.1 GBBI01002044 JAC16668.1 GEZM01086439 GEZM01086438 JAV59444.1 GBRD01016483 GBRD01016482 GDHC01012482 JAG49344.1 JAQ06147.1 KX602320 APP94029.1 KR075840 ALE20559.1 GECZ01007892 JAS61877.1 JN602375 KQ971338 AEO44887.1 KYB27979.1 GEZM01086441 GEZM01086437 JAV59437.1 UFQS01000789 UFQT01000789 SSX06891.1 SSX27235.1 GFDF01007701 JAV06383.1 GFDF01007164 JAV06920.1 GBYB01000831 JAG70598.1 GEDC01016201 JAS21097.1 GGLE01001962 MBY06088.1 GANO01003038 JAB56833.1 GGFK01002589 MBW35910.1 KU641425 MG680178 AQT40602.1 AWG41959.1 DQ440045 ABF18078.1 GFDL01001112 JAV33933.1 GFDL01001118 JAV33927.1 GALA01001182 JAA93670.1 GECZ01017425 JAS52344.1 GANP01011440 JAB73028.1 GBYB01007299 JAG77066.1 KY974307 ASW35107.1 KY923001 AUI80372.1 AAAB01008898 EGK97172.1 JR037830 AEY57993.1 GEDC01004317 JAS32981.1 KK122137 KFM81997.1 GEDC01019129 GEDC01000724 JAS18169.1 JAS36574.1 GBHO01029101 GBHO01025784 GBHO01017337 GBRD01016489 GBRD01016486 JAG14503.1 JAG17820.1 JAG26267.1 JAG49337.1 NEVH01007393 PNF35983.1 EF142066 ABO61882.1 GFAC01005508 JAT93680.1 GBBM01001268 JAC34150.1 GFAA01001287 JAU02148.1 GBBL01001221 JAC26099.1 GBBK01002606 JAC21876.1 IAAA01011464 LAA05187.1 AFFK01014267 GACK01004729 JAA60305.1 GFPF01008092 MAA19238.1 GEDV01001980 JAP86577.1 GDIP01007833 JAM95882.1 KK852559 KDR21360.1 GDAI01002144 JAI15459.1 GBGD01001716 JAC87173.1 GEFH01004028 JAP64553.1 GDIQ01184278 JAK67447.1 GECZ01021409 JAS48360.1 JR037831 AEY57994.1 GDIP01043960 JAM59755.1 GAKP01015698 JAC43254.1 GDIQ01225908 GDIP01112240 JAK25817.1 JAL91474.1 GDIP01120453 JAL83261.1 GFXV01007333 MBW19138.1 GGMR01018437 MBY31056.1 EU939764 ACH73246.1 GDIP01226788 JAI96613.1 GL732598 EFX72595.1 CH916371 EDV92330.1 GBHO01035195 GBHO01025781 GBHO01018235 GBRD01016485 GBRD01007877 JAG08409.1 JAG17823.1 JAG25369.1 JAG49341.1 GEBQ01004458 JAT35519.1 GAKP01015695 JAC43257.1 IACF01002588 LAB68236.1 GG666478 EEN66234.1 KT336322 ANZ22981.1 GAMC01007026 GAMC01007025 JAB99529.1 GBRD01016487 GBRD01007876 GDHC01015893 JAG49339.1 JAQ02736.1 GDHF01015274 JAI37040.1 KK119083 KFM74636.1 KU932409 APA34045.1 GEZM01086440 GEZM01086436 JAV59451.1 GGFL01006367 MBW70545.1 CH940651 EDW65721.1 KA646638 AFP61267.1 MUZQ01000007 OWK63988.1 LSYS01008075 OPJ69398.1 AADN05000813 AKCR02000007 PKK30500.1 ABQF01034770 ABQF01034771 ABQF01034772 ABQF01034773 KRF82353.1 CH933815 KRG07531.1 AHAT01020299 AHAT01020300 GDIQ01006531 JAN88206.1 KK107063 EZA61243.1 CP026256 AWP13254.1 AKHW03006231 KYO22436.1
Proteomes
Pfam
PF00069 Pkinase
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
A0A3S2NX33
A0A2A4JYU5
A0A0N7B1F3
S4P264
B0LBZ5
A0A2H1VZH2
+ More
A0A1B6C0U9 A0A1B6F6J0 T1H8J1 A0A194RV56 A0A0V0G765 A0A023F639 A0A1Y1KDH6 A0A0K8S854 A0A1L5YQK4 A0A0M4N2C8 A0A1B6GHH2 G3M3K4 A0A1Y1KKT3 A0A336KSE0 A0A1L8DIV1 A0A1L8DKQ2 A0A0C9R1V4 A0A1B6D5Y6 A0A2R5L9D4 U5EWA5 A0A2M4A5A8 A0A1S6LQ19 Q1HRP9 A0A1Q3G2A6 A0A1Q3G280 T1DE55 A0A1B6FQA7 V5HHR9 A0A0C9QU91 A0A286RXX4 A0A2I6BQ08 F5HLN9 V9IA57 A0A1B6E4X9 A0A087UXA8 A0A1B6EFD5 A0A0A9X6R6 A0A2J7R578 A9P773 A0A1E1X322 A0A023GMQ4 A0A1E1XS44 A0A023FX91 A0A023FIX7 A0A2L2YB23 T1IIH7 L7M8D6 A0A224YNQ5 A0A131Z5I0 A0A0P6C028 A0A067RL75 A0A0K8TM77 A0A069DSV4 A0A131XBN5 A0A0P5KKG9 A0A1B6FDU2 V9ICE0 A0A0P5ZK17 A0A034VLD1 A0A0P5I123 A0A0P5TXZ5 A0A2H8U153 A0A2S2PQD5 B5THN8 A0A0P4YJ16 E9H6Q6 B4JNN7 A0A0A9XG80 A0A1B6MI16 A0A034VMD5 A0A2P2I2X3 C3Y0G1 A0A1I9P1M5 W8C918 A0A0K8S849 A0A0K8VES1 A0A087UB97 A0A1I9WLM5 A0A1Y1KGF5 A0A2M4CZ54 B4M217 T1PFF3 A0A218VDR1 A0A1V4JB40 A0A3Q2TVV5 A0A2I0MLE2 H0ZT29 A0A0Q9WLE3 A0A0Q9XGA1 W5MJG8 A0A0P6IRZ3 A0A026WZI6 A0A2U9C9M7 A0A151MD28
A0A1B6C0U9 A0A1B6F6J0 T1H8J1 A0A194RV56 A0A0V0G765 A0A023F639 A0A1Y1KDH6 A0A0K8S854 A0A1L5YQK4 A0A0M4N2C8 A0A1B6GHH2 G3M3K4 A0A1Y1KKT3 A0A336KSE0 A0A1L8DIV1 A0A1L8DKQ2 A0A0C9R1V4 A0A1B6D5Y6 A0A2R5L9D4 U5EWA5 A0A2M4A5A8 A0A1S6LQ19 Q1HRP9 A0A1Q3G2A6 A0A1Q3G280 T1DE55 A0A1B6FQA7 V5HHR9 A0A0C9QU91 A0A286RXX4 A0A2I6BQ08 F5HLN9 V9IA57 A0A1B6E4X9 A0A087UXA8 A0A1B6EFD5 A0A0A9X6R6 A0A2J7R578 A9P773 A0A1E1X322 A0A023GMQ4 A0A1E1XS44 A0A023FX91 A0A023FIX7 A0A2L2YB23 T1IIH7 L7M8D6 A0A224YNQ5 A0A131Z5I0 A0A0P6C028 A0A067RL75 A0A0K8TM77 A0A069DSV4 A0A131XBN5 A0A0P5KKG9 A0A1B6FDU2 V9ICE0 A0A0P5ZK17 A0A034VLD1 A0A0P5I123 A0A0P5TXZ5 A0A2H8U153 A0A2S2PQD5 B5THN8 A0A0P4YJ16 E9H6Q6 B4JNN7 A0A0A9XG80 A0A1B6MI16 A0A034VMD5 A0A2P2I2X3 C3Y0G1 A0A1I9P1M5 W8C918 A0A0K8S849 A0A0K8VES1 A0A087UB97 A0A1I9WLM5 A0A1Y1KGF5 A0A2M4CZ54 B4M217 T1PFF3 A0A218VDR1 A0A1V4JB40 A0A3Q2TVV5 A0A2I0MLE2 H0ZT29 A0A0Q9WLE3 A0A0Q9XGA1 W5MJG8 A0A0P6IRZ3 A0A026WZI6 A0A2U9C9M7 A0A151MD28
PDB
4PTG
E-value=0,
Score=1653
Ontologies
PATHWAY
GO
GO:0005524
GO:0004674
GO:0004672
GO:1904781
GO:0001933
GO:0001954
GO:0031333
GO:0005886
GO:2000300
GO:0005813
GO:0043547
GO:0006983
GO:0001085
GO:0001837
GO:0071109
GO:0035176
GO:0032436
GO:0008285
GO:0098978
GO:0071625
GO:0002039
GO:0050321
GO:0030877
GO:0010822
GO:0002020
GO:0034452
GO:0008013
GO:0005634
GO:0070507
GO:0043066
GO:0021766
GO:0046827
GO:0008284
GO:1901215
GO:0031175
GO:0032091
GO:0018107
GO:0031334
GO:0046777
GO:0031625
GO:0070885
GO:0035556
GO:1901216
GO:0018105
GO:0034236
GO:0005977
GO:0051059
GO:0006468
GO:0016192
GO:0005839
GO:0051603
GO:0051028
GO:0005622
GO:0006810
GO:0003824
PANTHER
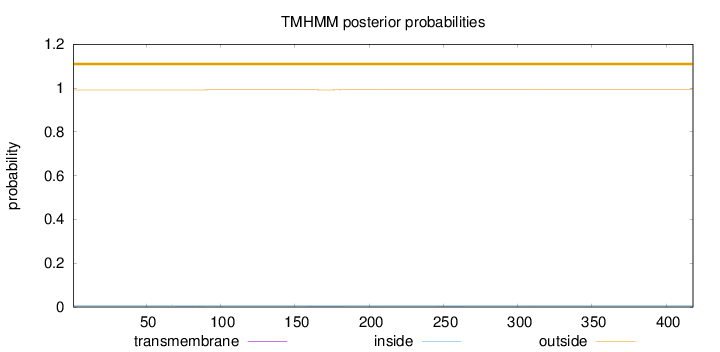
Topology
Length:
418
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02619
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00841
outside
1 - 418
Population Genetic Test Statistics
Pi
201.14805
Theta
171.021835
Tajima's D
0.67611
CLR
0.210568
CSRT
0.567871606419679
Interpretation
Possibly Positive selection
