Gene
KWMTBOMO10951 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008501
Annotation
PREDICTED:_serine/threonine-protein_phosphatase_PP1-beta_catalytic_subunit_[Papilio_polytes]
Full name
Serine/threonine-protein phosphatase
+ More
Serine/threonine-protein phosphatase PP1-beta catalytic subunit
Serine/threonine-protein phosphatase PP1-beta catalytic subunit
Location in the cell
Cytoplasmic Reliability : 2.612
Sequence
CDS
ATGGCTGACGAACTAAATGTTGACAGTCTAATTCACAGACTTTTGGAAGTTCGAGGATGCAGGCCTGGCAAGACTGTGCAAATGTCGGAGGCAGAGGTACGTGGGCTCTGCACAAAGTCCAGAGAAATATTCCTGCAGCAGCCCATATTACTGGAGTTGGAGGCACCACTCAAAATTTGTGGTGATATACATGGTCAGTACACAGATTTGCTACGTCTCTTTGAATATGGAGGATTTCCGCCAGAAGCCAATTACTTATTCCTTGGTGACTATGTAGATCGTGGTAAACAGTCACTGGAGACTATCTGTCTCCTATTGGCTTACAAAATAAAGTACCCTGAGAATTTCTTCCTACTCAGAGGCAATCATGAGTGTGCTAGCATAAACCGCATTTATGGTTTTTATGACGAGTGCAAACGCCGATACAATATAAAACTGTGGAAGACTTTCACGGATTGCTTTAACTGCTTGCCAATTGCTGCTATAATTGATGAGAAGATTTTCTGTTGCCATGGTGGTTTGTCTCCAGATTTACAGGGAATGGAACAGATCAGAAGAATAATGCGACCCACTGATGTTCCAGATACAGGATTGCTTTGTGACTTGCTGTGGTCAGATCCCGATAAGGATGTTCAAGGTTGGGGGGAAAACGATCGCGGCGTCTCGTTTACTTTCGGACCCGACGTCGTCAGTAAATTCCTCAACAGGCATGATTTAGATTTGATTTGCCGTGCTCACCAGGTGGTGGAAGATGGATATGAATTTTTCGCCAAACGAATGTTGGTTACTTTGTTCTCTGCACCCAACTACTGTGGAGAATTTGATAATGCTGGTGGAATGATGTCTGTTGATGAAACTCTAATGTGCTCCTTCCAGATTCTTAAACCATCTGAAAAAAAAGCAAAATACCAGTACAATGGTATAAACAGTGGAAGGCCAGCTACTCCTCAAAGGTCACCACCTAAGAAAAAATAA
Protein
MADELNVDSLIHRLLEVRGCRPGKTVQMSEAEVRGLCTKSREIFLQQPILLELEAPLKICGDIHGQYTDLLRLFEYGGFPPEANYLFLGDYVDRGKQSLETICLLLAYKIKYPENFFLLRGNHECASINRIYGFYDECKRRYNIKLWKTFTDCFNCLPIAAIIDEKIFCCHGGLSPDLQGMEQIRRIMRPTDVPDTGLLCDLLWSDPDKDVQGWGENDRGVSFTFGPDVVSKFLNRHDLDLICRAHQVVEDGYEFFAKRMLVTLFSAPNYCGEFDNAGGMMSVDETLMCSFQILKPSEKKAKYQYNGINSGRPATPQRSPPKKK
Summary
Description
Protein phosphatase that associates with over 200 regulatory proteins to form highly specific holoenzymes which dephosphorylate hundreds of biological targets. Protein phosphatase (PP1) is essential for cell division, it participates in the regulation of glycogen metabolism, muscle contractility and protein synthesis. Involved in regulation of ionic conductances and long-term synaptic plasticity (By similarity).
Catalytic Activity
H2O + O-phospho-L-seryl-[protein] = L-seryl-[protein] + phosphate
H2O + O-phospho-L-threonyl-[protein] = L-threonyl-[protein] + phosphate
H2O + O-phospho-L-threonyl-[protein] = L-threonyl-[protein] + phosphate
Cofactor
Mn(2+)
Similarity
Belongs to the PPP phosphatase family.
Belongs to the PPP phosphatase family. PP-1 subfamily.
Belongs to the PPP phosphatase family. PP-1 subfamily.
Keywords
Acetylation
Carbohydrate metabolism
Cell cycle
Cell division
Complete proteome
Cytoplasm
Glycogen metabolism
Hydrolase
Manganese
Metal-binding
Nucleus
Protein phosphatase
Reference proteome
Feature
chain Serine/threonine-protein phosphatase PP1-beta catalytic subunit
Uniprot
A0A194RQB4
A0A2H1WFH0
A0A194PN42
A0A212EZH9
A0A2W1BHD6
A0A067RHV6
+ More
A0A1B6H380 D2A1L2 A0A1B6LLR6 A0A1W4X671 A0A1B2M4Z2 E9IB51 A0A3M0KGY3 A0A0F8C489 A0A151WXC7 R4WDK9 E2C580 A0A0P4VMT2 K7INQ5 T1HE80 A0A158NB15 A0A026X1J8 A0A2J7RS99 A0A1B6EE98 A0A0A9X5V2 G1SEK1 A0A2K5CNF7 A0A224XJX0 A0A0V0G580 A0A087UKD3 A0A2K6QEJ2 A0A023FIW7 T1K2I3 A0A1B6DJX4 A0A210R013 A0A023GLJ1 A0A2R5LKD5 A0A224Z8Q5 A0A0K8RLZ2 A0A131YYT9 L7MB61 A0A023FWJ6 A0A293M3P6 A0A1Z5KWW7 A0A131XDT1 A0A2U9CUF0 A0A1L8G146 A0A3Q2HV97 A0A2I2V0N2 A0A1S3HZC0 A0A1E1XEM8 Q5I085 Q6GQL2 A0A2I0US19 A0A218UW58 A0A0S7KRU2 G1KIW6 A0A0B8RQ66 A0A1W7RDE9 T1E4U8 J3S0H7 U3FW09 A0A3Q0CW64 A0A250YFL0 A0A1J0N213 A0A1S3EYZ1 H0VAY6 A0A2K6C145 A0A2K6KHF2 L5KTG1 A0A2K5VEA0 K9IJA5 G1LET8 G1RZE5 A0A0D9RE27 G3SW90 A0A3Q2HEA5 A0A2U3V6V6 A0A340YHP4 A0A096P0A8 F7AJD1 A0A2Y9SAP0 A0A2K6QEJ4 A0A2U3YDE7 A0A2K5MSN5 A0A2Y9IEY6 A0A3Q7T4E9 F7BDA3 A0A1S2ZSM5 A0A3P4M0G5 M3Z2R4 A0A2K5RMM3 A0A2K6S1I8 A0A2Y9R8S4 A0A2J8VNA3 A0A2U3VJ48 A0A2Y9HP77 M3XDH6 A0A341A9E4 A0A3Q7U171
A0A1B6H380 D2A1L2 A0A1B6LLR6 A0A1W4X671 A0A1B2M4Z2 E9IB51 A0A3M0KGY3 A0A0F8C489 A0A151WXC7 R4WDK9 E2C580 A0A0P4VMT2 K7INQ5 T1HE80 A0A158NB15 A0A026X1J8 A0A2J7RS99 A0A1B6EE98 A0A0A9X5V2 G1SEK1 A0A2K5CNF7 A0A224XJX0 A0A0V0G580 A0A087UKD3 A0A2K6QEJ2 A0A023FIW7 T1K2I3 A0A1B6DJX4 A0A210R013 A0A023GLJ1 A0A2R5LKD5 A0A224Z8Q5 A0A0K8RLZ2 A0A131YYT9 L7MB61 A0A023FWJ6 A0A293M3P6 A0A1Z5KWW7 A0A131XDT1 A0A2U9CUF0 A0A1L8G146 A0A3Q2HV97 A0A2I2V0N2 A0A1S3HZC0 A0A1E1XEM8 Q5I085 Q6GQL2 A0A2I0US19 A0A218UW58 A0A0S7KRU2 G1KIW6 A0A0B8RQ66 A0A1W7RDE9 T1E4U8 J3S0H7 U3FW09 A0A3Q0CW64 A0A250YFL0 A0A1J0N213 A0A1S3EYZ1 H0VAY6 A0A2K6C145 A0A2K6KHF2 L5KTG1 A0A2K5VEA0 K9IJA5 G1LET8 G1RZE5 A0A0D9RE27 G3SW90 A0A3Q2HEA5 A0A2U3V6V6 A0A340YHP4 A0A096P0A8 F7AJD1 A0A2Y9SAP0 A0A2K6QEJ4 A0A2U3YDE7 A0A2K5MSN5 A0A2Y9IEY6 A0A3Q7T4E9 F7BDA3 A0A1S2ZSM5 A0A3P4M0G5 M3Z2R4 A0A2K5RMM3 A0A2K6S1I8 A0A2Y9R8S4 A0A2J8VNA3 A0A2U3VJ48 A0A2Y9HP77 M3XDH6 A0A341A9E4 A0A3Q7U171
EC Number
3.1.3.16
Pubmed
26354079
22118469
28756777
24845553
18362917
19820115
+ More
21282665 25835551 23691247 20798317 27129103 20075255 21347285 24508170 30249741 25401762 26823975 21993624 25362486 28812685 28797301 26830274 25576852 28528879 28049606 27762356 19892987 17975172 28503490 25476704 26358130 23758969 23025625 23915248 26319212 28087693 23258410 20010809 17495919 18464734
21282665 25835551 23691247 20798317 27129103 20075255 21347285 24508170 30249741 25401762 26823975 21993624 25362486 28812685 28797301 26830274 25576852 28528879 28049606 27762356 19892987 17975172 28503490 25476704 26358130 23758969 23025625 23915248 26319212 28087693 23258410 20010809 17495919 18464734
EMBL
KQ459765
KPJ20058.1
ODYU01008324
SOQ51813.1
KQ459604
KPI92540.1
+ More
AGBW02011299 OWR46861.1 KZ150127 PZC73104.1 KK852624 KDR19936.1 GECZ01000625 JAS69144.1 KQ971338 EFA02685.2 GEBQ01015468 JAT24509.1 KX467781 AOA60277.1 GL762111 EFZ22160.1 QRBI01000106 RMC12509.1 KQ042798 KKF10562.1 KQ982665 KYQ52564.1 AK417788 BAN21003.1 GL452767 EFN76901.1 GDKW01000725 JAI55870.1 ACPB03013340 ADTU01010746 KK107039 QOIP01000006 EZA61891.1 RLU21750.1 NEVH01000267 PNF43716.1 GEDC01018405 GEDC01001055 JAS18893.1 JAS36243.1 GBHO01028578 GBHO01028577 GBRD01014960 GDHC01000279 JAG15026.1 JAG15027.1 JAG50866.1 JAQ18350.1 AAGW02005738 AAGW02005739 AAGW02005740 GFTR01005082 JAW11344.1 GECL01003284 JAP02840.1 KK120226 KFM77822.1 GBBK01002930 JAC21552.1 CAEY01001363 GEDC01011304 GEDC01008183 JAS25994.1 JAS29115.1 NEDP02001069 OWF54353.1 GBBM01001733 JAC33685.1 GGLE01005858 MBY09984.1 GFPF01011726 MAA22872.1 GADI01001701 JAA72107.1 GEDV01004214 JAP84343.1 GACK01004611 JAA60423.1 GBBL01001446 JAC25874.1 GFWV01008876 MAA33605.1 GFJQ02007512 JAV99457.1 GEFH01003188 JAP65393.1 CP026262 AWP20257.1 CM004475 CM004474 OCT77504.1 OCT79525.1 AANG04003329 GFAC01001466 JAT97722.1 CR760533 BC088594 BC072730 KZ505646 PKU48820.1 MUZQ01000117 OWK57761.1 GBYX01200363 JAO79661.1 GBSH01002506 JAG66520.1 GDAY02002267 JAV49165.1 GAAZ01002311 JAA95632.1 JU175400 GBEX01003142 AFJ50924.1 JAI11418.1 GAEP01001490 GBEW01000949 JAB53331.1 JAI09416.1 GFFW01002361 JAV42427.1 KU871095 APD32937.1 AAKN02023477 AAKN02023478 KB030580 ELK14088.1 AQIA01018690 GABZ01006993 JAA46532.1 ACTA01012251 ACTA01020251 ADFV01011382 ADFV01011383 ADFV01011384 AQIB01033937 AQIB01033938 AQIB01033939 AQIB01033940 AQIB01033941 AQIB01033942 AQIB01033943 AQIB01033944 AHZZ02005103 AHZZ02005104 AHZZ02005105 CYRY02004269 VCW68787.1 AEYP01091144 AEYP01091145 AEYP01091146 AEYP01091147 NDHI03003415 PNJ59007.1
AGBW02011299 OWR46861.1 KZ150127 PZC73104.1 KK852624 KDR19936.1 GECZ01000625 JAS69144.1 KQ971338 EFA02685.2 GEBQ01015468 JAT24509.1 KX467781 AOA60277.1 GL762111 EFZ22160.1 QRBI01000106 RMC12509.1 KQ042798 KKF10562.1 KQ982665 KYQ52564.1 AK417788 BAN21003.1 GL452767 EFN76901.1 GDKW01000725 JAI55870.1 ACPB03013340 ADTU01010746 KK107039 QOIP01000006 EZA61891.1 RLU21750.1 NEVH01000267 PNF43716.1 GEDC01018405 GEDC01001055 JAS18893.1 JAS36243.1 GBHO01028578 GBHO01028577 GBRD01014960 GDHC01000279 JAG15026.1 JAG15027.1 JAG50866.1 JAQ18350.1 AAGW02005738 AAGW02005739 AAGW02005740 GFTR01005082 JAW11344.1 GECL01003284 JAP02840.1 KK120226 KFM77822.1 GBBK01002930 JAC21552.1 CAEY01001363 GEDC01011304 GEDC01008183 JAS25994.1 JAS29115.1 NEDP02001069 OWF54353.1 GBBM01001733 JAC33685.1 GGLE01005858 MBY09984.1 GFPF01011726 MAA22872.1 GADI01001701 JAA72107.1 GEDV01004214 JAP84343.1 GACK01004611 JAA60423.1 GBBL01001446 JAC25874.1 GFWV01008876 MAA33605.1 GFJQ02007512 JAV99457.1 GEFH01003188 JAP65393.1 CP026262 AWP20257.1 CM004475 CM004474 OCT77504.1 OCT79525.1 AANG04003329 GFAC01001466 JAT97722.1 CR760533 BC088594 BC072730 KZ505646 PKU48820.1 MUZQ01000117 OWK57761.1 GBYX01200363 JAO79661.1 GBSH01002506 JAG66520.1 GDAY02002267 JAV49165.1 GAAZ01002311 JAA95632.1 JU175400 GBEX01003142 AFJ50924.1 JAI11418.1 GAEP01001490 GBEW01000949 JAB53331.1 JAI09416.1 GFFW01002361 JAV42427.1 KU871095 APD32937.1 AAKN02023477 AAKN02023478 KB030580 ELK14088.1 AQIA01018690 GABZ01006993 JAA46532.1 ACTA01012251 ACTA01020251 ADFV01011382 ADFV01011383 ADFV01011384 AQIB01033937 AQIB01033938 AQIB01033939 AQIB01033940 AQIB01033941 AQIB01033942 AQIB01033943 AQIB01033944 AHZZ02005103 AHZZ02005104 AHZZ02005105 CYRY02004269 VCW68787.1 AEYP01091144 AEYP01091145 AEYP01091146 AEYP01091147 NDHI03003415 PNJ59007.1
Proteomes
UP000053240
UP000053268
UP000007151
UP000027135
UP000007266
UP000192223
+ More
UP000269221 UP000075809 UP000008237 UP000002358 UP000015103 UP000005205 UP000053097 UP000279307 UP000235965 UP000001811 UP000233020 UP000054359 UP000233200 UP000015104 UP000242188 UP000246464 UP000186698 UP000002281 UP000011712 UP000085678 UP000008143 UP000197619 UP000001646 UP000189706 UP000081671 UP000005447 UP000233120 UP000233180 UP000010552 UP000233100 UP000008912 UP000001073 UP000029965 UP000007646 UP000245320 UP000265300 UP000028761 UP000002280 UP000248484 UP000245341 UP000233060 UP000248482 UP000286640 UP000002279 UP000079721 UP000000715 UP000233040 UP000233220 UP000248480 UP000245340 UP000248481 UP000252040 UP000286642
UP000269221 UP000075809 UP000008237 UP000002358 UP000015103 UP000005205 UP000053097 UP000279307 UP000235965 UP000001811 UP000233020 UP000054359 UP000233200 UP000015104 UP000242188 UP000246464 UP000186698 UP000002281 UP000011712 UP000085678 UP000008143 UP000197619 UP000001646 UP000189706 UP000081671 UP000005447 UP000233120 UP000233180 UP000010552 UP000233100 UP000008912 UP000001073 UP000029965 UP000007646 UP000245320 UP000265300 UP000028761 UP000002280 UP000248484 UP000245341 UP000233060 UP000248482 UP000286640 UP000002279 UP000079721 UP000000715 UP000233040 UP000233220 UP000248480 UP000245340 UP000248481 UP000252040 UP000286642
Interpro
SUPFAM
SSF47473
SSF47473
Gene 3D
ProteinModelPortal
A0A194RQB4
A0A2H1WFH0
A0A194PN42
A0A212EZH9
A0A2W1BHD6
A0A067RHV6
+ More
A0A1B6H380 D2A1L2 A0A1B6LLR6 A0A1W4X671 A0A1B2M4Z2 E9IB51 A0A3M0KGY3 A0A0F8C489 A0A151WXC7 R4WDK9 E2C580 A0A0P4VMT2 K7INQ5 T1HE80 A0A158NB15 A0A026X1J8 A0A2J7RS99 A0A1B6EE98 A0A0A9X5V2 G1SEK1 A0A2K5CNF7 A0A224XJX0 A0A0V0G580 A0A087UKD3 A0A2K6QEJ2 A0A023FIW7 T1K2I3 A0A1B6DJX4 A0A210R013 A0A023GLJ1 A0A2R5LKD5 A0A224Z8Q5 A0A0K8RLZ2 A0A131YYT9 L7MB61 A0A023FWJ6 A0A293M3P6 A0A1Z5KWW7 A0A131XDT1 A0A2U9CUF0 A0A1L8G146 A0A3Q2HV97 A0A2I2V0N2 A0A1S3HZC0 A0A1E1XEM8 Q5I085 Q6GQL2 A0A2I0US19 A0A218UW58 A0A0S7KRU2 G1KIW6 A0A0B8RQ66 A0A1W7RDE9 T1E4U8 J3S0H7 U3FW09 A0A3Q0CW64 A0A250YFL0 A0A1J0N213 A0A1S3EYZ1 H0VAY6 A0A2K6C145 A0A2K6KHF2 L5KTG1 A0A2K5VEA0 K9IJA5 G1LET8 G1RZE5 A0A0D9RE27 G3SW90 A0A3Q2HEA5 A0A2U3V6V6 A0A340YHP4 A0A096P0A8 F7AJD1 A0A2Y9SAP0 A0A2K6QEJ4 A0A2U3YDE7 A0A2K5MSN5 A0A2Y9IEY6 A0A3Q7T4E9 F7BDA3 A0A1S2ZSM5 A0A3P4M0G5 M3Z2R4 A0A2K5RMM3 A0A2K6S1I8 A0A2Y9R8S4 A0A2J8VNA3 A0A2U3VJ48 A0A2Y9HP77 M3XDH6 A0A341A9E4 A0A3Q7U171
A0A1B6H380 D2A1L2 A0A1B6LLR6 A0A1W4X671 A0A1B2M4Z2 E9IB51 A0A3M0KGY3 A0A0F8C489 A0A151WXC7 R4WDK9 E2C580 A0A0P4VMT2 K7INQ5 T1HE80 A0A158NB15 A0A026X1J8 A0A2J7RS99 A0A1B6EE98 A0A0A9X5V2 G1SEK1 A0A2K5CNF7 A0A224XJX0 A0A0V0G580 A0A087UKD3 A0A2K6QEJ2 A0A023FIW7 T1K2I3 A0A1B6DJX4 A0A210R013 A0A023GLJ1 A0A2R5LKD5 A0A224Z8Q5 A0A0K8RLZ2 A0A131YYT9 L7MB61 A0A023FWJ6 A0A293M3P6 A0A1Z5KWW7 A0A131XDT1 A0A2U9CUF0 A0A1L8G146 A0A3Q2HV97 A0A2I2V0N2 A0A1S3HZC0 A0A1E1XEM8 Q5I085 Q6GQL2 A0A2I0US19 A0A218UW58 A0A0S7KRU2 G1KIW6 A0A0B8RQ66 A0A1W7RDE9 T1E4U8 J3S0H7 U3FW09 A0A3Q0CW64 A0A250YFL0 A0A1J0N213 A0A1S3EYZ1 H0VAY6 A0A2K6C145 A0A2K6KHF2 L5KTG1 A0A2K5VEA0 K9IJA5 G1LET8 G1RZE5 A0A0D9RE27 G3SW90 A0A3Q2HEA5 A0A2U3V6V6 A0A340YHP4 A0A096P0A8 F7AJD1 A0A2Y9SAP0 A0A2K6QEJ4 A0A2U3YDE7 A0A2K5MSN5 A0A2Y9IEY6 A0A3Q7T4E9 F7BDA3 A0A1S2ZSM5 A0A3P4M0G5 M3Z2R4 A0A2K5RMM3 A0A2K6S1I8 A0A2Y9R8S4 A0A2J8VNA3 A0A2U3VJ48 A0A2Y9HP77 M3XDH6 A0A341A9E4 A0A3Q7U171
PDB
1S70
E-value=0,
Score=1691
Ontologies
GO
GO:0004722
GO:0046872
GO:0000164
GO:0072357
GO:0005634
GO:0005737
GO:0004721
GO:0004620
GO:0016021
GO:0000784
GO:0005886
GO:0050115
GO:0042752
GO:0030155
GO:0005829
GO:0019901
GO:0051301
GO:0007049
GO:0032922
GO:0005977
GO:0043153
GO:0016787
GO:0008152
GO:0016491
GO:0016620
GO:0005839
GO:0051603
GO:0051028
GO:0005622
GO:0006810
GO:0003824
PANTHER
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleus
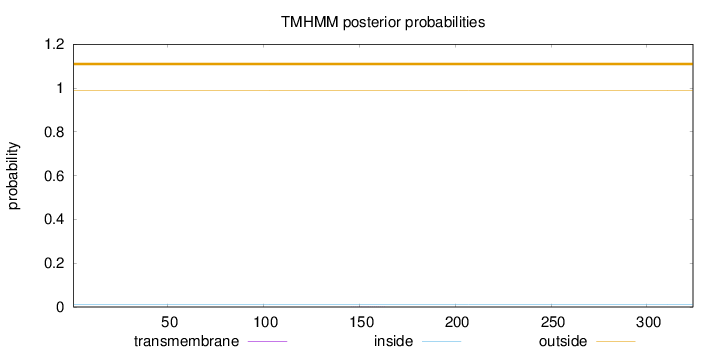
Length:
324
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00491999999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01174
outside
1 - 324
Population Genetic Test Statistics
Pi
220.682638
Theta
182.916556
Tajima's D
0.605235
CLR
0.030887
CSRT
0.543572821358932
Interpretation
Uncertain
