Gene
KWMTBOMO10946 Validated by peptides from experiments
Annotation
PREDICTED:_probable_methylmalonate-semialdehyde_dehydrogenase_[acylating]?_mitochondrial_[Plutella_xylostella]
Full name
Probable methylmalonate-semialdehyde dehydrogenase [acylating], mitochondrial
Location in the cell
Mitochondrial Reliability : 3.296
Sequence
CDS
ATGGCATCGGCGATCTTAAAGTTATTAAAATCTGAATCCCAGCTCGTACTGCGTCGCTCATATAGCAGCTCTGCTCCAACCACAAAGCTGTATATTGATGGACAATATGTAGATTCAAAAACTACCAATTGGATTGAGCTGACCAACCCAGCAACTAATGAAGTAATTGGCAGGGTGCCAGAGGCAACTCAGGATGAATTGACTTCCGCTTTGGATGCAGCCAAGAGGGCCTATAAGACATGGAGCAAAAGTACTGTTTTGACTCGTCAACAGCTAATGTTCAAATTTGCTCGGCTACTGAGAGAAAATCAGAGCAAATTGGCTGCTAAAATCACTGAAGAGCAAGGGAAAACTATAGCCGATGCTGAGGGCGATGTGCTTCGAGGAATTCAGTCAGTGGAGCATTGCTGTAGCATCACATCGCTTCAGCTCGGCGACAGCATCCAAAACATCGCCAAAGACATGGACACGCATAGCTACAAGGTGCCACTTGGAGTTGTTGGAGGTGTTGCAGCCTTCAATTTCCCGGTGATGATCCCACTGTGGATGTTCCCGCCAGCGCTGGTAACCGGAAACACGTGTATAATCAAGCCTTCTGAGCAGGATCCTGGAGCCACGCTCATGATGATGGAGCTCCTTCAGGAAGCCGGGGCTCCTCCGGGCGTTGTTAATATCATTCACGGGACACACGGTGCGGTGAACTTCATATGCGACCAGCCGGAGATCAAGGCCGTATCCTTCGTCGGTGGCGACGCCGCCGGAAAACACATCTACACTAGAGCTTCGGCTGCTGGCAAACGTGTCCAAAGCAACATGGGCGCCAAGAATCACGGGGTGATAATGCCGGACGCGAACAAGGAGCACACGCTCAACCAGCTCGCCGGCGCGGCCTTCGGAGCGGCCGGTCAACGGTGCATGGCGCTCAGCACTGCGGTCTTTGTCGGCAGTGCTAAGGAGTGGATCCCTGACCTGGTGAAGCGTGCGCAAGCTTTGAAGGTCAATGCGGGTCACGTGCCCGGCACCGACGTGGGGCCCGTCATCTCGGTAGCCGCCAAGGACAGAATCTTAAGGCTTGTGGAGTCAGGTGTAAAGGAAGGCGCTAAATTGGCTCTCGACGGTCGCGCGGTCAAAGTACCCGGATTCGAGAAAGGCAATTTCGTCGGGCCCACTATCATCACCGACGTGACGCCGAGCATGGAGTGTTACAAAGAGGAAATATTCGGGCCGGTTCTCGTCTGTCTGTTCGTTGAGACCCTCGACGAAGCGATTCAGATGATCAACTCAAACCCGTACGGGAATGGAACGGCTATTTTCACCACTAATGGTGCGACGGCTAGGAAGTTCTCCGCGGATATCGACGTCGGTCAAGTGGGCGTCAACGTGCCCATTCCCGTGCCGCTGTCGATGTTCTCGTTTTCGGGAACGCGCGGCAGTTTTCTGGGAACGAACCACTTCTGCGGGAAGCAGGGCCTCGACTTCTACACCGAACTGAAGACGGTCGTCTCGTTCTGGCGGGAGAGCGACGTGTCGCACGCGAAACCGGCCGTGTCTATGCCCACACAACAATAG
Protein
MASAILKLLKSESQLVLRRSYSSSAPTTKLYIDGQYVDSKTTNWIELTNPATNEVIGRVPEATQDELTSALDAAKRAYKTWSKSTVLTRQQLMFKFARLLRENQSKLAAKITEEQGKTIADAEGDVLRGIQSVEHCCSITSLQLGDSIQNIAKDMDTHSYKVPLGVVGGVAAFNFPVMIPLWMFPPALVTGNTCIIKPSEQDPGATLMMMELLQEAGAPPGVVNIIHGTHGAVNFICDQPEIKAVSFVGGDAAGKHIYTRASAAGKRVQSNMGAKNHGVIMPDANKEHTLNQLAGAAFGAAGQRCMALSTAVFVGSAKEWIPDLVKRAQALKVNAGHVPGTDVGPVISVAAKDRILRLVESGVKEGAKLALDGRAVKVPGFEKGNFVGPTIITDVTPSMECYKEEIFGPVLVCLFVETLDEAIQMINSNPYGNGTAIFTTNGATARKFSADIDVGQVGVNVPIPVPLSMFSFSGTRGSFLGTNHFCGKQGLDFYTELKTVVSFWRESDVSHAKPAVSMPTQQ
Summary
Description
Plays a role in valine and pyrimidine metabolism. Binds fatty acyl-CoA (By similarity).
Catalytic Activity
2-methyl-3-oxopropanoate + CoA + H2O + NAD(+) = H(+) + hydrogencarbonate + NADH + propanoyl-CoA
3-oxopropanoate + CoA + NAD(+) = acetyl-CoA + CO2 + NADH
3-oxopropanoate + CoA + NADP(+) = acetyl-CoA + CO2 + NADPH
3-oxopropanoate + CoA + NAD(+) = acetyl-CoA + CO2 + NADH
3-oxopropanoate + CoA + NADP(+) = acetyl-CoA + CO2 + NADPH
Subunit
Homotetramer.
Similarity
Belongs to the aldehyde dehydrogenase family.
Keywords
Complete proteome
Mitochondrion
NAD
Oxidoreductase
Reference proteome
Transit peptide
Alternative splicing
Feature
chain Probable methylmalonate-semialdehyde dehydrogenase [acylating], mitochondrial
splice variant In isoform A.
splice variant In isoform A.
Uniprot
A0A212FCD8
I4DMC8
A0A194PN47
A0A2W1BK47
A0A2A4J7U3
A0A194RRW7
+ More
S4PKI2 U5EUW1 A0A182IRL8 A0A1L8E2I7 Q17M80 A0A023ETJ6 A0A182XHX7 A0A182VWR2 A0A182VA95 A0A182TG60 A0A084VAP2 A0A182LEZ0 A0A182HSV9 Q7QC84 A0A1L8E2S9 A0A182N4V4 A0A1I8PJ02 A0A182MJI7 A0A182YHL4 A0A182P982 A0A182JZ74 A0A2J7PG61 A0A1L8EFN7 A0A182RGX2 A0A182QWJ8 T1E867 A0A2M4BK89 T1P9M5 A0A182FSB4 A0A182SQR9 A0A0L0CHF9 A0A2M3YYR2 W5JA91 A0A2M4A0N5 A0A2M3ZFI7 A0A0M4EU10 A0A2M4A229 B0WSQ7 A0A1Q3FKZ1 A0A1W4V8D5 B4MAJ9 B3MSF3 B4I926 B3P971 B4PX02 A0A1L8EBR3 Q7KW39 I0B1N8 B4L668 B4R7E7 B4NCQ5 B4JM08 Q29HB2 A0A034WNG4 A0A224YTE8 A0A3B0KRJ4 A0A131YMU9 A0A0K8V8N8 A0A1J1HJY4 A0A067QPJ5 Q7KW39-2 A0A0A1X2P7 W8BE83 A0A336M0R0 B4GY04 A0A0N8EQ03 A0A0P5EQI7 A0A0P5VKV8 A0A0P5MF92 A0A0P4ZVA4 A0A1Z5KWX0 A0A0P5MNL3 A0A0P5AQT1 A0A0P4ZN91 A0A0P5PFK7 A0A0P5LBJ1 A0A0P5EKC6 A0A0P5E2J8 V5IF88 A0A0P5AHP3 A0A0P5M625 A0A0P5JA01 A0A0P5BFD6 A0A1A9W8R5 A0A0P5M2I5 A0A0P6E113 A0A0P4ZU12 T1IIJ5 E9GHJ1 A0A0P6A917 E0VIN4 A0A131Y2Q8 A0A0P4ZUH7 A0A1L8EAL5
S4PKI2 U5EUW1 A0A182IRL8 A0A1L8E2I7 Q17M80 A0A023ETJ6 A0A182XHX7 A0A182VWR2 A0A182VA95 A0A182TG60 A0A084VAP2 A0A182LEZ0 A0A182HSV9 Q7QC84 A0A1L8E2S9 A0A182N4V4 A0A1I8PJ02 A0A182MJI7 A0A182YHL4 A0A182P982 A0A182JZ74 A0A2J7PG61 A0A1L8EFN7 A0A182RGX2 A0A182QWJ8 T1E867 A0A2M4BK89 T1P9M5 A0A182FSB4 A0A182SQR9 A0A0L0CHF9 A0A2M3YYR2 W5JA91 A0A2M4A0N5 A0A2M3ZFI7 A0A0M4EU10 A0A2M4A229 B0WSQ7 A0A1Q3FKZ1 A0A1W4V8D5 B4MAJ9 B3MSF3 B4I926 B3P971 B4PX02 A0A1L8EBR3 Q7KW39 I0B1N8 B4L668 B4R7E7 B4NCQ5 B4JM08 Q29HB2 A0A034WNG4 A0A224YTE8 A0A3B0KRJ4 A0A131YMU9 A0A0K8V8N8 A0A1J1HJY4 A0A067QPJ5 Q7KW39-2 A0A0A1X2P7 W8BE83 A0A336M0R0 B4GY04 A0A0N8EQ03 A0A0P5EQI7 A0A0P5VKV8 A0A0P5MF92 A0A0P4ZVA4 A0A1Z5KWX0 A0A0P5MNL3 A0A0P5AQT1 A0A0P4ZN91 A0A0P5PFK7 A0A0P5LBJ1 A0A0P5EKC6 A0A0P5E2J8 V5IF88 A0A0P5AHP3 A0A0P5M625 A0A0P5JA01 A0A0P5BFD6 A0A1A9W8R5 A0A0P5M2I5 A0A0P6E113 A0A0P4ZU12 T1IIJ5 E9GHJ1 A0A0P6A917 E0VIN4 A0A131Y2Q8 A0A0P4ZUH7 A0A1L8EAL5
Pubmed
EMBL
AGBW02009208
OWR51377.1
AK402446
BAM19068.1
KQ459604
KPI92545.1
+ More
KZ150127 PZC73100.1 NWSH01002487 PCG68197.1 KQ459765 KPJ20065.1 GAIX01004480 JAA88080.1 GANO01001278 JAB58593.1 GFDF01001134 JAV12950.1 CH477208 EAT47764.1 JXUM01034772 JXUM01034773 JXUM01092490 GAPW01000960 KQ563990 KQ561036 JAC12638.1 KXJ72967.1 KXJ79938.1 ATLV01004135 KE524190 KFB35036.1 APCN01000368 AAAB01008859 GFDF01001128 JAV12956.1 AXCM01001791 NEVH01025635 PNF15320.1 GFDG01001390 JAV17409.1 AXCN02000010 GAMD01003033 JAA98557.1 GGFJ01004326 MBW53467.1 KA644633 AFP59262.1 JRES01000394 KNC31681.1 GGFM01000649 MBW21400.1 ADMH02001952 ETN60333.1 GGFK01001046 MBW34367.1 GGFM01006512 MBW27263.1 CP012528 ALC48485.1 GGFK01001469 MBW34790.1 DS232075 EDS34016.1 GFDL01006798 JAV28247.1 CH940655 EDW66258.1 CH902622 EDV34708.1 CH480825 EDW43707.1 CH954183 EDV45367.2 CM000162 EDX00788.2 GFDG01002744 JAV16055.1 AE014298 AL009147 AY069284 AAF45510.2 CAA15632.1 BT133399 AFH55498.1 CH933812 EDW05864.1 CM000366 EDX16758.1 CH964239 EDW82614.1 CH916371 EDV91769.1 CH379064 EAL31846.1 GAKP01003055 JAC55897.1 GFPF01009710 MAA20856.1 OUUW01000019 SPP89269.1 GEDV01008797 JAP79760.1 GDHF01017354 JAI34960.1 CVRI01000006 CRK87858.1 KK853778 KDQ78911.1 GBXI01009267 JAD05025.1 GAMC01009603 JAB96952.1 UFQT01000386 SSX23814.1 CH479196 EDW27631.1 GDIQ01005244 JAN89493.1 GDIP01156503 JAJ66899.1 GDIP01113353 GDIP01043262 LRGB01002580 JAL90361.1 KZS06852.1 GDIQ01160416 JAK91309.1 GDIP01210935 JAJ12467.1 GFJQ02007410 JAV99559.1 GDIQ01176610 JAK75115.1 GDIP01195783 JAJ27619.1 GDIP01210936 JAJ12466.1 GDIQ01235199 GDIQ01129281 JAL22445.1 GDIQ01176609 JAK75116.1 GDIP01158588 JAJ64814.1 GDIP01147346 GDIP01147344 JAJ76058.1 GANP01008862 JAB75606.1 GDIP01199038 JAJ24364.1 GDIQ01184208 JAK67517.1 GDIQ01204903 JAK46822.1 GDIP01185435 JAJ37967.1 GDIQ01185954 JAK65771.1 GDIQ01081145 JAN13592.1 GDIP01208541 JAJ14861.1 JH430174 GL732545 EFX80854.1 GDIP01032597 JAM71118.1 DS235201 EEB13240.1 GEFM01003034 JAP72762.1 GDIP01208542 JAJ14860.1 GFDG01003137 JAV15662.1
KZ150127 PZC73100.1 NWSH01002487 PCG68197.1 KQ459765 KPJ20065.1 GAIX01004480 JAA88080.1 GANO01001278 JAB58593.1 GFDF01001134 JAV12950.1 CH477208 EAT47764.1 JXUM01034772 JXUM01034773 JXUM01092490 GAPW01000960 KQ563990 KQ561036 JAC12638.1 KXJ72967.1 KXJ79938.1 ATLV01004135 KE524190 KFB35036.1 APCN01000368 AAAB01008859 GFDF01001128 JAV12956.1 AXCM01001791 NEVH01025635 PNF15320.1 GFDG01001390 JAV17409.1 AXCN02000010 GAMD01003033 JAA98557.1 GGFJ01004326 MBW53467.1 KA644633 AFP59262.1 JRES01000394 KNC31681.1 GGFM01000649 MBW21400.1 ADMH02001952 ETN60333.1 GGFK01001046 MBW34367.1 GGFM01006512 MBW27263.1 CP012528 ALC48485.1 GGFK01001469 MBW34790.1 DS232075 EDS34016.1 GFDL01006798 JAV28247.1 CH940655 EDW66258.1 CH902622 EDV34708.1 CH480825 EDW43707.1 CH954183 EDV45367.2 CM000162 EDX00788.2 GFDG01002744 JAV16055.1 AE014298 AL009147 AY069284 AAF45510.2 CAA15632.1 BT133399 AFH55498.1 CH933812 EDW05864.1 CM000366 EDX16758.1 CH964239 EDW82614.1 CH916371 EDV91769.1 CH379064 EAL31846.1 GAKP01003055 JAC55897.1 GFPF01009710 MAA20856.1 OUUW01000019 SPP89269.1 GEDV01008797 JAP79760.1 GDHF01017354 JAI34960.1 CVRI01000006 CRK87858.1 KK853778 KDQ78911.1 GBXI01009267 JAD05025.1 GAMC01009603 JAB96952.1 UFQT01000386 SSX23814.1 CH479196 EDW27631.1 GDIQ01005244 JAN89493.1 GDIP01156503 JAJ66899.1 GDIP01113353 GDIP01043262 LRGB01002580 JAL90361.1 KZS06852.1 GDIQ01160416 JAK91309.1 GDIP01210935 JAJ12467.1 GFJQ02007410 JAV99559.1 GDIQ01176610 JAK75115.1 GDIP01195783 JAJ27619.1 GDIP01210936 JAJ12466.1 GDIQ01235199 GDIQ01129281 JAL22445.1 GDIQ01176609 JAK75116.1 GDIP01158588 JAJ64814.1 GDIP01147346 GDIP01147344 JAJ76058.1 GANP01008862 JAB75606.1 GDIP01199038 JAJ24364.1 GDIQ01184208 JAK67517.1 GDIQ01204903 JAK46822.1 GDIP01185435 JAJ37967.1 GDIQ01185954 JAK65771.1 GDIQ01081145 JAN13592.1 GDIP01208541 JAJ14861.1 JH430174 GL732545 EFX80854.1 GDIP01032597 JAM71118.1 DS235201 EEB13240.1 GEFM01003034 JAP72762.1 GDIP01208542 JAJ14860.1 GFDG01003137 JAV15662.1
Proteomes
UP000007151
UP000053268
UP000218220
UP000053240
UP000075880
UP000008820
+ More
UP000069940 UP000249989 UP000076407 UP000075920 UP000075903 UP000075902 UP000030765 UP000075882 UP000075840 UP000007062 UP000075884 UP000095300 UP000075883 UP000076408 UP000075885 UP000075881 UP000235965 UP000075900 UP000075886 UP000095301 UP000069272 UP000075901 UP000037069 UP000000673 UP000092553 UP000002320 UP000192221 UP000008792 UP000007801 UP000001292 UP000008711 UP000002282 UP000000803 UP000009192 UP000000304 UP000007798 UP000001070 UP000001819 UP000268350 UP000183832 UP000027135 UP000008744 UP000076858 UP000091820 UP000000305 UP000009046
UP000069940 UP000249989 UP000076407 UP000075920 UP000075903 UP000075902 UP000030765 UP000075882 UP000075840 UP000007062 UP000075884 UP000095300 UP000075883 UP000076408 UP000075885 UP000075881 UP000235965 UP000075900 UP000075886 UP000095301 UP000069272 UP000075901 UP000037069 UP000000673 UP000092553 UP000002320 UP000192221 UP000008792 UP000007801 UP000001292 UP000008711 UP000002282 UP000000803 UP000009192 UP000000304 UP000007798 UP000001070 UP000001819 UP000268350 UP000183832 UP000027135 UP000008744 UP000076858 UP000091820 UP000000305 UP000009046
Pfam
Interpro
IPR016162
Ald_DH_N
+ More
IPR015590 Aldehyde_DH_dom
IPR016161 Ald_DH/histidinol_DH
IPR010061 MeMal-semiAld_DH
IPR016160 Ald_DH_CS_CYS
IPR016163 Ald_DH_C
IPR004947 DNase_II
IPR002109 Glutaredoxin
IPR033658 GRX_PICOT-like
IPR004480 Monothiol_GRX-rel
IPR036249 Thioredoxin-like_sf
IPR001180 CNH_dom
IPR032914 Vam6/VPS39/TRAP1
IPR019453 VPS39/TGF_beta_rcpt-assoc_2
IPR019452 VPS39/TGF_beta_rcpt-assoc_1
IPR000547 Clathrin_H-chain/VPS_repeat
IPR015590 Aldehyde_DH_dom
IPR016161 Ald_DH/histidinol_DH
IPR010061 MeMal-semiAld_DH
IPR016160 Ald_DH_CS_CYS
IPR016163 Ald_DH_C
IPR004947 DNase_II
IPR002109 Glutaredoxin
IPR033658 GRX_PICOT-like
IPR004480 Monothiol_GRX-rel
IPR036249 Thioredoxin-like_sf
IPR001180 CNH_dom
IPR032914 Vam6/VPS39/TRAP1
IPR019453 VPS39/TGF_beta_rcpt-assoc_2
IPR019452 VPS39/TGF_beta_rcpt-assoc_1
IPR000547 Clathrin_H-chain/VPS_repeat
Gene 3D
ProteinModelPortal
A0A212FCD8
I4DMC8
A0A194PN47
A0A2W1BK47
A0A2A4J7U3
A0A194RRW7
+ More
S4PKI2 U5EUW1 A0A182IRL8 A0A1L8E2I7 Q17M80 A0A023ETJ6 A0A182XHX7 A0A182VWR2 A0A182VA95 A0A182TG60 A0A084VAP2 A0A182LEZ0 A0A182HSV9 Q7QC84 A0A1L8E2S9 A0A182N4V4 A0A1I8PJ02 A0A182MJI7 A0A182YHL4 A0A182P982 A0A182JZ74 A0A2J7PG61 A0A1L8EFN7 A0A182RGX2 A0A182QWJ8 T1E867 A0A2M4BK89 T1P9M5 A0A182FSB4 A0A182SQR9 A0A0L0CHF9 A0A2M3YYR2 W5JA91 A0A2M4A0N5 A0A2M3ZFI7 A0A0M4EU10 A0A2M4A229 B0WSQ7 A0A1Q3FKZ1 A0A1W4V8D5 B4MAJ9 B3MSF3 B4I926 B3P971 B4PX02 A0A1L8EBR3 Q7KW39 I0B1N8 B4L668 B4R7E7 B4NCQ5 B4JM08 Q29HB2 A0A034WNG4 A0A224YTE8 A0A3B0KRJ4 A0A131YMU9 A0A0K8V8N8 A0A1J1HJY4 A0A067QPJ5 Q7KW39-2 A0A0A1X2P7 W8BE83 A0A336M0R0 B4GY04 A0A0N8EQ03 A0A0P5EQI7 A0A0P5VKV8 A0A0P5MF92 A0A0P4ZVA4 A0A1Z5KWX0 A0A0P5MNL3 A0A0P5AQT1 A0A0P4ZN91 A0A0P5PFK7 A0A0P5LBJ1 A0A0P5EKC6 A0A0P5E2J8 V5IF88 A0A0P5AHP3 A0A0P5M625 A0A0P5JA01 A0A0P5BFD6 A0A1A9W8R5 A0A0P5M2I5 A0A0P6E113 A0A0P4ZU12 T1IIJ5 E9GHJ1 A0A0P6A917 E0VIN4 A0A131Y2Q8 A0A0P4ZUH7 A0A1L8EAL5
S4PKI2 U5EUW1 A0A182IRL8 A0A1L8E2I7 Q17M80 A0A023ETJ6 A0A182XHX7 A0A182VWR2 A0A182VA95 A0A182TG60 A0A084VAP2 A0A182LEZ0 A0A182HSV9 Q7QC84 A0A1L8E2S9 A0A182N4V4 A0A1I8PJ02 A0A182MJI7 A0A182YHL4 A0A182P982 A0A182JZ74 A0A2J7PG61 A0A1L8EFN7 A0A182RGX2 A0A182QWJ8 T1E867 A0A2M4BK89 T1P9M5 A0A182FSB4 A0A182SQR9 A0A0L0CHF9 A0A2M3YYR2 W5JA91 A0A2M4A0N5 A0A2M3ZFI7 A0A0M4EU10 A0A2M4A229 B0WSQ7 A0A1Q3FKZ1 A0A1W4V8D5 B4MAJ9 B3MSF3 B4I926 B3P971 B4PX02 A0A1L8EBR3 Q7KW39 I0B1N8 B4L668 B4R7E7 B4NCQ5 B4JM08 Q29HB2 A0A034WNG4 A0A224YTE8 A0A3B0KRJ4 A0A131YMU9 A0A0K8V8N8 A0A1J1HJY4 A0A067QPJ5 Q7KW39-2 A0A0A1X2P7 W8BE83 A0A336M0R0 B4GY04 A0A0N8EQ03 A0A0P5EQI7 A0A0P5VKV8 A0A0P5MF92 A0A0P4ZVA4 A0A1Z5KWX0 A0A0P5MNL3 A0A0P5AQT1 A0A0P4ZN91 A0A0P5PFK7 A0A0P5LBJ1 A0A0P5EKC6 A0A0P5E2J8 V5IF88 A0A0P5AHP3 A0A0P5M625 A0A0P5JA01 A0A0P5BFD6 A0A1A9W8R5 A0A0P5M2I5 A0A0P6E113 A0A0P4ZU12 T1IIJ5 E9GHJ1 A0A0P6A917 E0VIN4 A0A131Y2Q8 A0A0P4ZUH7 A0A1L8EAL5
PDB
5TJR
E-value=2.16967e-118,
Score=1090
Ontologies
PATHWAY
00280
Valine, leucine and isoleucine degradation - Bombyx mori (domestic silkworm)
00410 beta-Alanine metabolism - Bombyx mori (domestic silkworm)
00562 Inositol phosphate metabolism - Bombyx mori (domestic silkworm)
00640 Propanoate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
00410 beta-Alanine metabolism - Bombyx mori (domestic silkworm)
00562 Inositol phosphate metabolism - Bombyx mori (domestic silkworm)
00640 Propanoate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
GO
Topology
Subcellular location
Mitochondrion
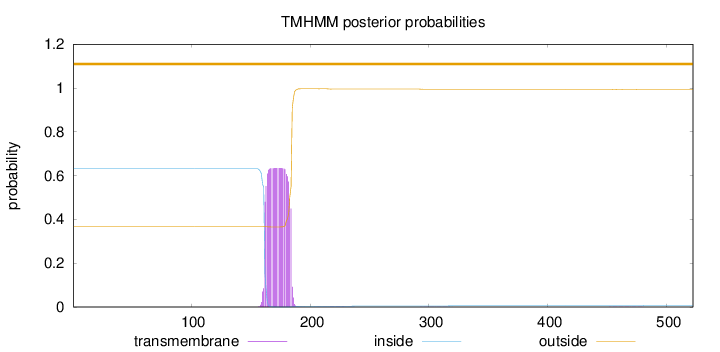
Length:
522
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
14.28517
Exp number, first 60 AAs:
0.00036
Total prob of N-in:
0.63159
outside
1 - 522
Population Genetic Test Statistics
Pi
151.854746
Theta
160.65364
Tajima's D
0.191707
CLR
2.262863
CSRT
0.430728463576821
Interpretation
Uncertain
