Gene
KWMTBOMO10936 Validated by peptides from experiments
Annotation
hypothetical_protein_KGM_19166_[Danaus_plexippus]
Full name
Proteasome subunit beta
Location in the cell
Extracellular Reliability : 1.091
Sequence
CDS
ATGTCAAATATAAACTTACAATGTCTCTTAGGAATACAGTGCAACGACTTTGTTATGATAGCAGCCGATCAGAGCAACAGCCACAGCATTATGATCATGAAAGACGATGAAGAAAAGATCTATAAGATATCTGATAGATTAGTAATGGGTGTCATTGGTGACTCAGGAGACACAAATCAATTTGCTGAATATATTGCAAAAAATATTCAGTTGTATAAAATGCGTAATGGGTATGAATTGGGGCCCTCAGCTGCAGCCAGTTTCACACGTCGTAATCTCGCAGAGTATCTTAGGAGCAGTACTCCCTACTTTGTAAATGTGCTCATGGGTGGTTATGATAAAGAAAATGGACCTGAACTGTATTTTATGGACTATTTAGCATCAAGCGTGAAGGTACCATTTGCAGCTCATGGTTATGGTGGATATCTAAGTTTGAGTATTATGGATCGTTACCATAAAAAAGATGCTACAGAAACAGAGGCATATGACATCCTTAAGAAGTGTGTTCAAGAAGTTCACAAAAGATTGTTTGTTAGTCTCCCCAATTTCCAAGTGACTGTTGTAAACCGTGATGGGATTAAAGTGCTGCCCACCATTAATTCTGCATCTCTTCAATAA
Protein
MSNINLQCLLGIQCNDFVMIAADQSNSHSIMIMKDDEEKIYKISDRLVMGVIGDSGDTNQFAEYIAKNIQLYKMRNGYELGPSAAASFTRRNLAEYLRSSTPYFVNVLMGGYDKENGPELYFMDYLASSVKVPFAAHGYGGYLSLSIMDRYHKKDATETEAYDILKKCVQEVHKRLFVSLPNFQVTVVNRDGIKVLPTINSASLQ
Summary
Catalytic Activity
Cleavage of peptide bonds with very broad specificity.
Subunit
The 26S proteasome consists of a 20S proteasome core and two 19S regulatory subunits.
Similarity
Belongs to the peptidase T1B family.
Feature
chain Proteasome subunit beta
Uniprot
A0A3S2LY20
A0A194RRH8
A0A194PIK4
S4PBF7
A0A2W1BRR4
A0A2A4K3Q1
+ More
A0A2H1VJ26 A0A1E1WQZ4 A0A212FCC9 A0A2J7PHZ7 A0A154PNC2 A0A087ZWB2 E2BY51 I4DP24 A0A2A3EMN4 A0A0L7QVH1 A0A067R3P7 A0A026X561 A0A0P5Z5F8 A0A0P5WU15 A0A0P5M4S4 A0A0P5QF09 A0A0P5QKR7 A0A0N8DTF6 A0A0P5A981 A0A0N8C7G3 A0A232EMC0 K7J9Q1 A0A151IML7 A0A0P6ILM8 A0A0P5YD11 E9IB35 A0A151XHU4 F4X4Y2 A0A151HY76 A0A195ESA4 A0A158NVX6 A0A310SF94 A0A0N8BRN9 A0A0N8AXZ3 E9HF20 A0A0P5Q8R2 A0A0P5NJY9 A0A0N0BEU3 A0A1Y1NKF7 A0A0P5YST0 A0A0P5THG0 A0A0P5NWN9 A2IAA2 D6WPM7 R9TFV9 A0A293MNJ5 A0A1S6GL72 V5GZQ3 E0VWC5 A0A2H1V2W2 J3JU85 A0A2R5LE76 A0A1Z5LEK4 A0A1W4WMT1 A0A1S3J3M5 A0A0P5TVH2 B0W1D3 U5EV66 A0A1Q3FDP7 A0A0A9XMJ8 A0A224YS56 A0A224YQ14 A0A023FU65 A0A023ELL5 Q17IE3 A0A0K8TSM5 A0A1B6E889 A0A131YV56 A0A0K8S7L4 A0A2W1BKD8 A0A1S4EGJ4 A0A336JWZ1 A0A1E1X5E0 A0A182SCD9 A0A182GQ96 K1PDC6 A0A182Y980 A0A3S2L821 A0A182R4A9 A0A182VU49 R7THR9 A0A0P5NGD4 Q7PWV4 A0A023EIC0 A0A2A4JXW0 Q6GQ40 A0A182PMF4 A0A0K8R840 A0A182IJ98 A0A084VYF7 A0A1S4H053 A0A182VD31 A0A182UCY6 A0A182XDH7
A0A2H1VJ26 A0A1E1WQZ4 A0A212FCC9 A0A2J7PHZ7 A0A154PNC2 A0A087ZWB2 E2BY51 I4DP24 A0A2A3EMN4 A0A0L7QVH1 A0A067R3P7 A0A026X561 A0A0P5Z5F8 A0A0P5WU15 A0A0P5M4S4 A0A0P5QF09 A0A0P5QKR7 A0A0N8DTF6 A0A0P5A981 A0A0N8C7G3 A0A232EMC0 K7J9Q1 A0A151IML7 A0A0P6ILM8 A0A0P5YD11 E9IB35 A0A151XHU4 F4X4Y2 A0A151HY76 A0A195ESA4 A0A158NVX6 A0A310SF94 A0A0N8BRN9 A0A0N8AXZ3 E9HF20 A0A0P5Q8R2 A0A0P5NJY9 A0A0N0BEU3 A0A1Y1NKF7 A0A0P5YST0 A0A0P5THG0 A0A0P5NWN9 A2IAA2 D6WPM7 R9TFV9 A0A293MNJ5 A0A1S6GL72 V5GZQ3 E0VWC5 A0A2H1V2W2 J3JU85 A0A2R5LE76 A0A1Z5LEK4 A0A1W4WMT1 A0A1S3J3M5 A0A0P5TVH2 B0W1D3 U5EV66 A0A1Q3FDP7 A0A0A9XMJ8 A0A224YS56 A0A224YQ14 A0A023FU65 A0A023ELL5 Q17IE3 A0A0K8TSM5 A0A1B6E889 A0A131YV56 A0A0K8S7L4 A0A2W1BKD8 A0A1S4EGJ4 A0A336JWZ1 A0A1E1X5E0 A0A182SCD9 A0A182GQ96 K1PDC6 A0A182Y980 A0A3S2L821 A0A182R4A9 A0A182VU49 R7THR9 A0A0P5NGD4 Q7PWV4 A0A023EIC0 A0A2A4JXW0 Q6GQ40 A0A182PMF4 A0A0K8R840 A0A182IJ98 A0A084VYF7 A0A1S4H053 A0A182VD31 A0A182UCY6 A0A182XDH7
EC Number
3.4.25.1
Pubmed
26354079
23622113
28756777
22118469
20798317
22651552
+ More
24845553 24508170 30249741 28648823 20075255 21282665 21719571 21347285 21292972 28004739 17437641 18362917 19820115 28043022 20566863 22516182 23537049 28528879 25401762 26823975 28797301 24945155 26483478 17510324 26369729 26830274 28503490 22992520 25244985 23254933 12364791 27762356 24438588
24845553 24508170 30249741 28648823 20075255 21282665 21719571 21347285 21292972 28004739 17437641 18362917 19820115 28043022 20566863 22516182 23537049 28528879 25401762 26823975 28797301 24945155 26483478 17510324 26369729 26830274 28503490 22992520 25244985 23254933 12364791 27762356 24438588
EMBL
RSAL01001425
RSAL01000123
RVE40795.1
RVE46634.1
KQ459765
KPJ20072.1
+ More
KQ459604 KPI92554.1 GAIX01005932 JAA86628.1 KZ149920 PZC77769.1 NWSH01000165 PCG78887.1 ODYU01002848 SOQ40839.1 GDQN01001803 JAT89251.1 AGBW02009208 OWR51367.1 NEVH01025130 PNF15952.1 KQ434978 KZC12964.1 GL451420 EFN79330.1 AK403258 BAM19664.1 KZ288215 PBC32532.1 KQ414727 KOC62491.1 KK852779 KDR16684.1 KK107020 QOIP01000006 EZA62569.1 RLU21138.1 GDIP01061421 JAM42294.1 GDIP01081769 JAM21946.1 GDIQ01184930 GDIQ01180584 GDIQ01046076 JAK66795.1 GDIQ01121404 JAL30322.1 GDIQ01112850 JAL38876.1 GDIP01002599 LRGB01003216 JAN01117.1 KZS03717.1 GDIP01205751 JAJ17651.1 GDIQ01107355 JAL44371.1 NNAY01003382 OXU19505.1 AAZX01001624 KQ977033 KYN06198.1 GDIQ01019830 JAN74907.1 GDIP01059470 JAM44245.1 GL762106 EFZ22229.1 KQ982129 KYQ59858.1 GL888679 EGI58555.1 KQ976730 KYM76394.1 KQ981993 KYN31130.1 ADTU01027584 KQ769068 OAD52975.1 GDIQ01151547 JAL00179.1 GDIQ01231915 JAK19810.1 GL732633 EFX69663.1 GDIQ01139143 JAL12583.1 GDIQ01141148 JAL10578.1 KQ435821 KOX72411.1 GEZM01000453 GEZM01000452 JAV98413.1 GDIP01055077 JAM48638.1 GDIP01127812 JAL75902.1 GDIQ01136649 JAL15077.1 EF179416 ABM55422.1 KQ971354 EFA06200.1 KC989882 KC989883 AGN29655.1 GFWV01022540 MAA47267.1 KY314165 AQS22599.1 GALX01002473 JAB65993.1 DS235818 EEB17681.1 ODYU01000215 SOQ34614.1 APGK01054913 BT126798 KB741253 KB632340 AEE61760.1 ENN71815.1 ERL92957.1 GGLE01003714 MBY07840.1 GFJQ02001134 JAW05836.1 GDIP01121413 JAL82301.1 DS231821 EDS45003.1 GANO01002000 JAB57871.1 GFDL01009406 JAV25639.1 GBHO01021652 GDHC01019431 JAG21952.1 JAP99197.1 GFPF01005504 MAA16650.1 GFPF01005505 MAA16651.1 GBBL01001959 JAC25361.1 JXUM01108243 GAPW01004449 KQ565213 JAC09149.1 KXJ71212.1 CH477240 EAT46434.1 GDAI01000480 JAI17123.1 GEDC01003147 JAS34151.1 GEDV01006152 JAP82405.1 GBRD01016548 JAG49278.1 KZ150214 PZC72143.1 UFQS01000002 UFQT01000002 SSW96721.1 SSX17108.1 GFAC01004698 JAT94490.1 JXUM01015684 JXUM01015685 KQ560437 KXJ82419.1 JH815987 EKC21877.1 RSAL01000095 RVE47821.1 AMQN01012817 KB309774 ELT93338.1 GDIQ01149196 JAL02530.1 AAAB01008984 EAA14834.3 GAPW01004450 JAC09148.1 NWSH01000405 PCG76649.1 BC072908 BC106262 CM004468 AAH72908.1 AAI06263.1 OCT94763.1 GADI01006476 JAA67332.1 ATLV01018364 KE525231 KFB43001.1
KQ459604 KPI92554.1 GAIX01005932 JAA86628.1 KZ149920 PZC77769.1 NWSH01000165 PCG78887.1 ODYU01002848 SOQ40839.1 GDQN01001803 JAT89251.1 AGBW02009208 OWR51367.1 NEVH01025130 PNF15952.1 KQ434978 KZC12964.1 GL451420 EFN79330.1 AK403258 BAM19664.1 KZ288215 PBC32532.1 KQ414727 KOC62491.1 KK852779 KDR16684.1 KK107020 QOIP01000006 EZA62569.1 RLU21138.1 GDIP01061421 JAM42294.1 GDIP01081769 JAM21946.1 GDIQ01184930 GDIQ01180584 GDIQ01046076 JAK66795.1 GDIQ01121404 JAL30322.1 GDIQ01112850 JAL38876.1 GDIP01002599 LRGB01003216 JAN01117.1 KZS03717.1 GDIP01205751 JAJ17651.1 GDIQ01107355 JAL44371.1 NNAY01003382 OXU19505.1 AAZX01001624 KQ977033 KYN06198.1 GDIQ01019830 JAN74907.1 GDIP01059470 JAM44245.1 GL762106 EFZ22229.1 KQ982129 KYQ59858.1 GL888679 EGI58555.1 KQ976730 KYM76394.1 KQ981993 KYN31130.1 ADTU01027584 KQ769068 OAD52975.1 GDIQ01151547 JAL00179.1 GDIQ01231915 JAK19810.1 GL732633 EFX69663.1 GDIQ01139143 JAL12583.1 GDIQ01141148 JAL10578.1 KQ435821 KOX72411.1 GEZM01000453 GEZM01000452 JAV98413.1 GDIP01055077 JAM48638.1 GDIP01127812 JAL75902.1 GDIQ01136649 JAL15077.1 EF179416 ABM55422.1 KQ971354 EFA06200.1 KC989882 KC989883 AGN29655.1 GFWV01022540 MAA47267.1 KY314165 AQS22599.1 GALX01002473 JAB65993.1 DS235818 EEB17681.1 ODYU01000215 SOQ34614.1 APGK01054913 BT126798 KB741253 KB632340 AEE61760.1 ENN71815.1 ERL92957.1 GGLE01003714 MBY07840.1 GFJQ02001134 JAW05836.1 GDIP01121413 JAL82301.1 DS231821 EDS45003.1 GANO01002000 JAB57871.1 GFDL01009406 JAV25639.1 GBHO01021652 GDHC01019431 JAG21952.1 JAP99197.1 GFPF01005504 MAA16650.1 GFPF01005505 MAA16651.1 GBBL01001959 JAC25361.1 JXUM01108243 GAPW01004449 KQ565213 JAC09149.1 KXJ71212.1 CH477240 EAT46434.1 GDAI01000480 JAI17123.1 GEDC01003147 JAS34151.1 GEDV01006152 JAP82405.1 GBRD01016548 JAG49278.1 KZ150214 PZC72143.1 UFQS01000002 UFQT01000002 SSW96721.1 SSX17108.1 GFAC01004698 JAT94490.1 JXUM01015684 JXUM01015685 KQ560437 KXJ82419.1 JH815987 EKC21877.1 RSAL01000095 RVE47821.1 AMQN01012817 KB309774 ELT93338.1 GDIQ01149196 JAL02530.1 AAAB01008984 EAA14834.3 GAPW01004450 JAC09148.1 NWSH01000405 PCG76649.1 BC072908 BC106262 CM004468 AAH72908.1 AAI06263.1 OCT94763.1 GADI01006476 JAA67332.1 ATLV01018364 KE525231 KFB43001.1
Proteomes
UP000283053
UP000053240
UP000053268
UP000218220
UP000007151
UP000235965
+ More
UP000076502 UP000005203 UP000008237 UP000242457 UP000053825 UP000027135 UP000053097 UP000279307 UP000076858 UP000215335 UP000002358 UP000078542 UP000075809 UP000007755 UP000078540 UP000078541 UP000005205 UP000000305 UP000053105 UP000007266 UP000009046 UP000019118 UP000030742 UP000192223 UP000085678 UP000002320 UP000069940 UP000249989 UP000008820 UP000079169 UP000075901 UP000005408 UP000076408 UP000075900 UP000075920 UP000014760 UP000007062 UP000186698 UP000075885 UP000075880 UP000030765 UP000075903 UP000075902 UP000076407
UP000076502 UP000005203 UP000008237 UP000242457 UP000053825 UP000027135 UP000053097 UP000279307 UP000076858 UP000215335 UP000002358 UP000078542 UP000075809 UP000007755 UP000078540 UP000078541 UP000005205 UP000000305 UP000053105 UP000007266 UP000009046 UP000019118 UP000030742 UP000192223 UP000085678 UP000002320 UP000069940 UP000249989 UP000008820 UP000079169 UP000075901 UP000005408 UP000076408 UP000075900 UP000075920 UP000014760 UP000007062 UP000186698 UP000075885 UP000075880 UP000030765 UP000075903 UP000075902 UP000076407
PRIDE
Pfam
PF00227 Proteasome
Interpro
SUPFAM
SSF56235
SSF56235
Gene 3D
ProteinModelPortal
A0A3S2LY20
A0A194RRH8
A0A194PIK4
S4PBF7
A0A2W1BRR4
A0A2A4K3Q1
+ More
A0A2H1VJ26 A0A1E1WQZ4 A0A212FCC9 A0A2J7PHZ7 A0A154PNC2 A0A087ZWB2 E2BY51 I4DP24 A0A2A3EMN4 A0A0L7QVH1 A0A067R3P7 A0A026X561 A0A0P5Z5F8 A0A0P5WU15 A0A0P5M4S4 A0A0P5QF09 A0A0P5QKR7 A0A0N8DTF6 A0A0P5A981 A0A0N8C7G3 A0A232EMC0 K7J9Q1 A0A151IML7 A0A0P6ILM8 A0A0P5YD11 E9IB35 A0A151XHU4 F4X4Y2 A0A151HY76 A0A195ESA4 A0A158NVX6 A0A310SF94 A0A0N8BRN9 A0A0N8AXZ3 E9HF20 A0A0P5Q8R2 A0A0P5NJY9 A0A0N0BEU3 A0A1Y1NKF7 A0A0P5YST0 A0A0P5THG0 A0A0P5NWN9 A2IAA2 D6WPM7 R9TFV9 A0A293MNJ5 A0A1S6GL72 V5GZQ3 E0VWC5 A0A2H1V2W2 J3JU85 A0A2R5LE76 A0A1Z5LEK4 A0A1W4WMT1 A0A1S3J3M5 A0A0P5TVH2 B0W1D3 U5EV66 A0A1Q3FDP7 A0A0A9XMJ8 A0A224YS56 A0A224YQ14 A0A023FU65 A0A023ELL5 Q17IE3 A0A0K8TSM5 A0A1B6E889 A0A131YV56 A0A0K8S7L4 A0A2W1BKD8 A0A1S4EGJ4 A0A336JWZ1 A0A1E1X5E0 A0A182SCD9 A0A182GQ96 K1PDC6 A0A182Y980 A0A3S2L821 A0A182R4A9 A0A182VU49 R7THR9 A0A0P5NGD4 Q7PWV4 A0A023EIC0 A0A2A4JXW0 Q6GQ40 A0A182PMF4 A0A0K8R840 A0A182IJ98 A0A084VYF7 A0A1S4H053 A0A182VD31 A0A182UCY6 A0A182XDH7
A0A2H1VJ26 A0A1E1WQZ4 A0A212FCC9 A0A2J7PHZ7 A0A154PNC2 A0A087ZWB2 E2BY51 I4DP24 A0A2A3EMN4 A0A0L7QVH1 A0A067R3P7 A0A026X561 A0A0P5Z5F8 A0A0P5WU15 A0A0P5M4S4 A0A0P5QF09 A0A0P5QKR7 A0A0N8DTF6 A0A0P5A981 A0A0N8C7G3 A0A232EMC0 K7J9Q1 A0A151IML7 A0A0P6ILM8 A0A0P5YD11 E9IB35 A0A151XHU4 F4X4Y2 A0A151HY76 A0A195ESA4 A0A158NVX6 A0A310SF94 A0A0N8BRN9 A0A0N8AXZ3 E9HF20 A0A0P5Q8R2 A0A0P5NJY9 A0A0N0BEU3 A0A1Y1NKF7 A0A0P5YST0 A0A0P5THG0 A0A0P5NWN9 A2IAA2 D6WPM7 R9TFV9 A0A293MNJ5 A0A1S6GL72 V5GZQ3 E0VWC5 A0A2H1V2W2 J3JU85 A0A2R5LE76 A0A1Z5LEK4 A0A1W4WMT1 A0A1S3J3M5 A0A0P5TVH2 B0W1D3 U5EV66 A0A1Q3FDP7 A0A0A9XMJ8 A0A224YS56 A0A224YQ14 A0A023FU65 A0A023ELL5 Q17IE3 A0A0K8TSM5 A0A1B6E889 A0A131YV56 A0A0K8S7L4 A0A2W1BKD8 A0A1S4EGJ4 A0A336JWZ1 A0A1E1X5E0 A0A182SCD9 A0A182GQ96 K1PDC6 A0A182Y980 A0A3S2L821 A0A182R4A9 A0A182VU49 R7THR9 A0A0P5NGD4 Q7PWV4 A0A023EIC0 A0A2A4JXW0 Q6GQ40 A0A182PMF4 A0A0K8R840 A0A182IJ98 A0A084VYF7 A0A1S4H053 A0A182VD31 A0A182UCY6 A0A182XDH7
PDB
3UNH
E-value=9.99321e-57,
Score=553
Ontologies
GO
PANTHER
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleus
Length:
205
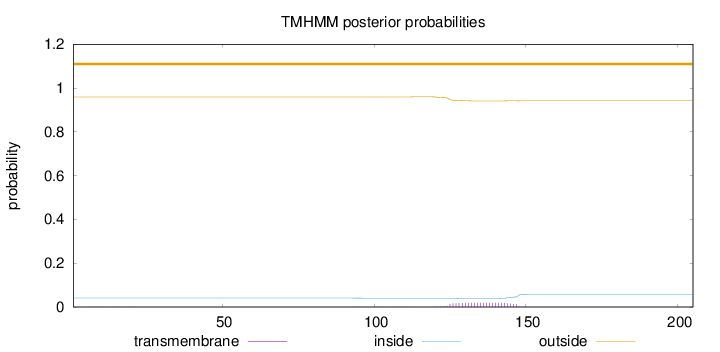
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.44315
Exp number, first 60 AAs:
0.00324
Total prob of N-in:
0.04112
outside
1 - 205
Population Genetic Test Statistics
Pi
127.048439
Theta
142.613292
Tajima's D
-0.417432
CLR
11.458177
CSRT
0.260236988150592
Interpretation
Uncertain
