Gene
KWMTBOMO10935
Pre Gene Modal
BGIBMGA008497
Annotation
PREDICTED:_transcription_initiation_factor_TFIID_subunit_8_[Amyelois_transitella]
Full name
Transcription initiation factor TFIID subunit 8
Alternative Name
Protein prodos
Location in the cell
Nuclear Reliability : 2.787
Sequence
CDS
ATGTCAGAAAAAACTGAAACTACATCTGATGCTCGTCGACGTATCTTAAACATTGCAGTTTCTACGGTATTATTGGAGACCGGTTTCGATGCTGCTGATAAAGTTTCGTTGGAAACTTTGACTGAAATGCTTCAATGTTTTTTTGCAGAAGTTGGGAATTCGGCGAAAGGATTTTGTGAGCTATCTGGACGGGTGGAGCCAGTGTTGGGTGATGTTGTAATGGCACTTATTAACATGGGTATAAGCTTACAGGGTATTGAACAATATGCTGCTCGTCCAAATCGGCATGTAATTCAACCTCCTCAGCAAGCTCCTGCACCTCGCACCCCAGCTATGTTGTCAGCTGGCTCTAAAGCTAAACCAGCACCACATATACCTTTTCATTTGCCTCCATTACCAGATCCACATGCATACATCAGAACACCTACTCACAAGCAACCAGTAACTGAATATGAAGCCATTCGAGAGAAGGCAGCAACCCAAAAAAGAGACATTGAACGTGCTTTGACTAAATTCTTAGCAAAGACAAGTGAAACCCATAATTTATTTAATACAGAAGACAATCAGGTTTTTCCATTAATCGCCTGTAAGCCTACGTTTCCAGCTTATTTGCCCTGTCTTTTACCAACGGACCAAGTCTTTGATTTTGATGAATTAGAATATCATTTTCAAGTCGCTAACAGAACTGAAGACTTGCCTGCTGATAAAAAAGATCAATCAGACAATGAAGGCGAAAATGGCGCTGACAATGATAATGCAAATATTTCACAATCTGAAAATCAAGAAAGTGCATTAACATCCAGTCCAGAGAGAAATAAATCTGGTGGATAG
Protein
MSEKTETTSDARRRILNIAVSTVLLETGFDAADKVSLETLTEMLQCFFAEVGNSAKGFCELSGRVEPVLGDVVMALINMGISLQGIEQYAARPNRHVIQPPQQAPAPRTPAMLSAGSKAKPAPHIPFHLPPLPDPHAYIRTPTHKQPVTEYEAIREKAATQKRDIERALTKFLAKTSETHNLFNTEDNQVFPLIACKPTFPAYLPCLLPTDQVFDFDELEYHFQVANRTEDLPADKKDQSDNEGENGADNDNANISQSENQESALTSSPERNKSGG
Summary
Description
TFIID is a multimeric protein complex that plays a central role in mediating promoter responses to various activators and repressors.
Subunit
Belongs to the TFIID complex which is composed of TATA binding protein (Tbp) and a number of TBP-associated factors (TAFs). Histone fold interacts with N-terminus of Taf10b.
Similarity
Belongs to the TAF8 family.
Keywords
Complete proteome
Nucleus
Phosphoprotein
Reference proteome
Transcription
Transcription regulation
Feature
chain Transcription initiation factor TFIID subunit 8
Uniprot
A0A194RV79
A0A194PHM7
A0A2H1VJ30
A0A2A4K4Q1
A0A2W1BY76
A0A212FCC5
+ More
I4DRV0 A0A3S2NBE9 S4PSZ0 H9JG50 A0A1L8DI39 A0A034WFA8 W8C8M9 A0A0K8WDZ2 A0A0K8WCS7 A0A0L0BUE7 Q174K2 A0A1B0FAF2 A0A182GWU5 A0A1A9Z1P9 A0A1A9X8T9 A0A1B0C3B7 A0A084WIY5 A0A1A9WTD5 A0A1I8N819 A0A336KVU8 A0A1A9VRK7 A0A182IRZ1 A0A2M3ZJ73 A0A2M3ZJ63 A0A1Q3F9X3 A0A2M4BUC4 A0A2M4BUB4 W5JGV7 A0A182R170 A0A2M4ARF4 B0WBV2 A0A1I8PSF3 A0A1I8PSK3 A0A182JZV1 A0A182X066 A0A182KT88 A0A182HPL2 A0A1I8PSI1 A0A182P1F7 A0A182YC88 A0A182M742 A0A182FTY4 A0A182N3R5 B4L2H9 B4M7V9 A0A0L7QQR6 Q7PZV2 A0A154PH42 T1PHY5 B4Q2U6 A0A2A3EE76 A0A088ASJ7 B3NTA7 U5EJQ4 B4I6N7 A0A0M4EUQ8 A0A1W4V0R5 H1UUC4 Q9VWY6 D3TQ88 B3MXU6 E2ACF0 A0A182RAI0 A0A195BBZ0 A0A158NMU2 A0A0M9ABS9 A0A195EXV9 B4JXF0 Q29H69 A0A151WUP8 A0A0J7NRY3 B4H082 A0A151JBE8 F4WTB0 A0A3L8DKT6 A0A026WM59 B4NQ20 A0A195C7L0 A0A2P8YHZ8 K7IMW3 E2BTN5 A0A182TEM4 A0A182VFQ5 A0A2J7PX97 A0A1J1HHU9 E0VGZ1 A0A067QYI5 A0A1W4XLI9 B0WBV1 A0A069DQF3 A0A232FF83 A0A023F9C6 A0A224XVQ6
I4DRV0 A0A3S2NBE9 S4PSZ0 H9JG50 A0A1L8DI39 A0A034WFA8 W8C8M9 A0A0K8WDZ2 A0A0K8WCS7 A0A0L0BUE7 Q174K2 A0A1B0FAF2 A0A182GWU5 A0A1A9Z1P9 A0A1A9X8T9 A0A1B0C3B7 A0A084WIY5 A0A1A9WTD5 A0A1I8N819 A0A336KVU8 A0A1A9VRK7 A0A182IRZ1 A0A2M3ZJ73 A0A2M3ZJ63 A0A1Q3F9X3 A0A2M4BUC4 A0A2M4BUB4 W5JGV7 A0A182R170 A0A2M4ARF4 B0WBV2 A0A1I8PSF3 A0A1I8PSK3 A0A182JZV1 A0A182X066 A0A182KT88 A0A182HPL2 A0A1I8PSI1 A0A182P1F7 A0A182YC88 A0A182M742 A0A182FTY4 A0A182N3R5 B4L2H9 B4M7V9 A0A0L7QQR6 Q7PZV2 A0A154PH42 T1PHY5 B4Q2U6 A0A2A3EE76 A0A088ASJ7 B3NTA7 U5EJQ4 B4I6N7 A0A0M4EUQ8 A0A1W4V0R5 H1UUC4 Q9VWY6 D3TQ88 B3MXU6 E2ACF0 A0A182RAI0 A0A195BBZ0 A0A158NMU2 A0A0M9ABS9 A0A195EXV9 B4JXF0 Q29H69 A0A151WUP8 A0A0J7NRY3 B4H082 A0A151JBE8 F4WTB0 A0A3L8DKT6 A0A026WM59 B4NQ20 A0A195C7L0 A0A2P8YHZ8 K7IMW3 E2BTN5 A0A182TEM4 A0A182VFQ5 A0A2J7PX97 A0A1J1HHU9 E0VGZ1 A0A067QYI5 A0A1W4XLI9 B0WBV1 A0A069DQF3 A0A232FF83 A0A023F9C6 A0A224XVQ6
Pubmed
26354079
28756777
22118469
22651552
23622113
19121390
+ More
25348373 24495485 26108605 17510324 26483478 24438588 25315136 20920257 23761445 20966253 25244985 17994087 12364791 17550304 10731132 12537572 12537569 11134347 18327897 20353571 20798317 21347285 15632085 21719571 30249741 24508170 29403074 20075255 20566863 24845553 26334808 28648823 25474469
25348373 24495485 26108605 17510324 26483478 24438588 25315136 20920257 23761445 20966253 25244985 17994087 12364791 17550304 10731132 12537572 12537569 11134347 18327897 20353571 20798317 21347285 15632085 21719571 30249741 24508170 29403074 20075255 20566863 24845553 26334808 28648823 25474469
EMBL
KQ459765
KPJ20071.1
KQ459604
KPI92553.1
ODYU01002848
SOQ40840.1
+ More
NWSH01000165 PCG78888.1 KZ149920 PZC77770.1 AGBW02009208 OWR51368.1 AK405279 BAM20640.1 RSAL01000123 RVE46633.1 GAIX01012953 JAA79607.1 BABH01001989 GFDF01007945 JAV06139.1 GAKP01005563 JAC53389.1 GAMC01002894 JAC03662.1 GDHF01010422 GDHF01003012 JAI41892.1 JAI49302.1 GDHF01004293 GDHF01003326 JAI48021.1 JAI48988.1 JRES01001305 KNC23695.1 CH477409 EAT41503.1 CCAG010015202 JXUM01094154 KQ564107 KXJ72779.1 JXJN01024882 ATLV01023954 KE525347 KFB50179.1 UFQS01001076 UFQT01001076 SSX08857.1 SSX28768.1 GGFM01007876 MBW28627.1 GGFM01007866 MBW28617.1 GFDL01010689 JAV24356.1 GGFJ01007462 MBW56603.1 GGFJ01007463 MBW56604.1 ADMH02001571 ETN62039.1 AXCN02001100 GGFK01009857 MBW43178.1 DS231882 EDS42882.1 APCN01003265 AXCM01006018 CH933810 EDW06855.1 CH940653 EDW62876.2 KQ414785 KOC60975.1 AAAB01008986 EAA00622.4 KQ434902 KZC11175.1 KA648427 AFP63056.1 CM000162 EDX02680.1 KZ288271 PBC29774.1 CH954180 EDV46558.1 GANO01002156 JAB57715.1 CH480823 EDW55985.1 CP012528 ALC48860.1 BT133008 AEX66194.1 Y15513 AE014298 AY071604 AAF48800.1 AAL49226.1 CAA75667.1 EZ423590 ADD19866.1 CH902630 EDV38561.1 GL438494 EFN68889.1 KQ976529 KYM81737.1 ADTU01020809 KQ435710 KOX79919.1 KQ981940 KYN32732.1 CH916376 EDV95426.1 CH379064 EAL31889.2 KQ982721 KYQ51649.1 LBMM01002163 KMQ95195.1 CH479200 EDW29677.1 KQ979147 KYN22446.1 GL888334 EGI62573.1 QOIP01000007 RLU20519.1 KK107152 EZA57137.1 CH964291 EDW86245.1 KQ978231 KYM96151.1 PYGN01000580 PSN43879.1 GL450467 EFN80950.1 NEVH01020861 PNF20950.1 CVRI01000004 CRK87616.1 DS235154 EEB12647.1 KK852822 KDR15520.1 EDS42881.1 GBGD01002591 JAC86298.1 NNAY01000352 OXU28977.1 GBBI01000610 JAC18102.1 GFTR01004233 JAW12193.1
NWSH01000165 PCG78888.1 KZ149920 PZC77770.1 AGBW02009208 OWR51368.1 AK405279 BAM20640.1 RSAL01000123 RVE46633.1 GAIX01012953 JAA79607.1 BABH01001989 GFDF01007945 JAV06139.1 GAKP01005563 JAC53389.1 GAMC01002894 JAC03662.1 GDHF01010422 GDHF01003012 JAI41892.1 JAI49302.1 GDHF01004293 GDHF01003326 JAI48021.1 JAI48988.1 JRES01001305 KNC23695.1 CH477409 EAT41503.1 CCAG010015202 JXUM01094154 KQ564107 KXJ72779.1 JXJN01024882 ATLV01023954 KE525347 KFB50179.1 UFQS01001076 UFQT01001076 SSX08857.1 SSX28768.1 GGFM01007876 MBW28627.1 GGFM01007866 MBW28617.1 GFDL01010689 JAV24356.1 GGFJ01007462 MBW56603.1 GGFJ01007463 MBW56604.1 ADMH02001571 ETN62039.1 AXCN02001100 GGFK01009857 MBW43178.1 DS231882 EDS42882.1 APCN01003265 AXCM01006018 CH933810 EDW06855.1 CH940653 EDW62876.2 KQ414785 KOC60975.1 AAAB01008986 EAA00622.4 KQ434902 KZC11175.1 KA648427 AFP63056.1 CM000162 EDX02680.1 KZ288271 PBC29774.1 CH954180 EDV46558.1 GANO01002156 JAB57715.1 CH480823 EDW55985.1 CP012528 ALC48860.1 BT133008 AEX66194.1 Y15513 AE014298 AY071604 AAF48800.1 AAL49226.1 CAA75667.1 EZ423590 ADD19866.1 CH902630 EDV38561.1 GL438494 EFN68889.1 KQ976529 KYM81737.1 ADTU01020809 KQ435710 KOX79919.1 KQ981940 KYN32732.1 CH916376 EDV95426.1 CH379064 EAL31889.2 KQ982721 KYQ51649.1 LBMM01002163 KMQ95195.1 CH479200 EDW29677.1 KQ979147 KYN22446.1 GL888334 EGI62573.1 QOIP01000007 RLU20519.1 KK107152 EZA57137.1 CH964291 EDW86245.1 KQ978231 KYM96151.1 PYGN01000580 PSN43879.1 GL450467 EFN80950.1 NEVH01020861 PNF20950.1 CVRI01000004 CRK87616.1 DS235154 EEB12647.1 KK852822 KDR15520.1 EDS42881.1 GBGD01002591 JAC86298.1 NNAY01000352 OXU28977.1 GBBI01000610 JAC18102.1 GFTR01004233 JAW12193.1
Proteomes
UP000053240
UP000053268
UP000218220
UP000007151
UP000283053
UP000005204
+ More
UP000037069 UP000008820 UP000092444 UP000069940 UP000249989 UP000092445 UP000092443 UP000092460 UP000030765 UP000091820 UP000095301 UP000078200 UP000075880 UP000000673 UP000075886 UP000002320 UP000095300 UP000075881 UP000076407 UP000075882 UP000075840 UP000075885 UP000076408 UP000075883 UP000069272 UP000075884 UP000009192 UP000008792 UP000053825 UP000007062 UP000076502 UP000002282 UP000242457 UP000005203 UP000008711 UP000001292 UP000092553 UP000192221 UP000000803 UP000007801 UP000000311 UP000075900 UP000078540 UP000005205 UP000053105 UP000078541 UP000001070 UP000001819 UP000075809 UP000036403 UP000008744 UP000078492 UP000007755 UP000279307 UP000053097 UP000007798 UP000078542 UP000245037 UP000002358 UP000008237 UP000075902 UP000075903 UP000235965 UP000183832 UP000009046 UP000027135 UP000192223 UP000215335
UP000037069 UP000008820 UP000092444 UP000069940 UP000249989 UP000092445 UP000092443 UP000092460 UP000030765 UP000091820 UP000095301 UP000078200 UP000075880 UP000000673 UP000075886 UP000002320 UP000095300 UP000075881 UP000076407 UP000075882 UP000075840 UP000075885 UP000076408 UP000075883 UP000069272 UP000075884 UP000009192 UP000008792 UP000053825 UP000007062 UP000076502 UP000002282 UP000242457 UP000005203 UP000008711 UP000001292 UP000092553 UP000192221 UP000000803 UP000007801 UP000000311 UP000075900 UP000078540 UP000005205 UP000053105 UP000078541 UP000001070 UP000001819 UP000075809 UP000036403 UP000008744 UP000078492 UP000007755 UP000279307 UP000053097 UP000007798 UP000078542 UP000245037 UP000002358 UP000008237 UP000075902 UP000075903 UP000235965 UP000183832 UP000009046 UP000027135 UP000192223 UP000215335
Interpro
IPR037818
TAF8
+ More
IPR006565 BTP
IPR009072 Histone-fold
IPR019473 TFIID_su8_C
IPR006638 Elp3/MiaB/NifB
IPR005839 Methylthiotransferase
IPR006463 MiaB_methiolase
IPR002792 TRAM_dom
IPR013848 Methylthiotransferase_N
IPR023404 rSAM_horseshoe
IPR038135 Methylthiotransferase_N_sf
IPR020612 Methylthiotransferase_CS
IPR007197 rSAM
IPR006565 BTP
IPR009072 Histone-fold
IPR019473 TFIID_su8_C
IPR006638 Elp3/MiaB/NifB
IPR005839 Methylthiotransferase
IPR006463 MiaB_methiolase
IPR002792 TRAM_dom
IPR013848 Methylthiotransferase_N
IPR023404 rSAM_horseshoe
IPR038135 Methylthiotransferase_N_sf
IPR020612 Methylthiotransferase_CS
IPR007197 rSAM
SUPFAM
SSF47113
SSF47113
Gene 3D
CDD
ProteinModelPortal
A0A194RV79
A0A194PHM7
A0A2H1VJ30
A0A2A4K4Q1
A0A2W1BY76
A0A212FCC5
+ More
I4DRV0 A0A3S2NBE9 S4PSZ0 H9JG50 A0A1L8DI39 A0A034WFA8 W8C8M9 A0A0K8WDZ2 A0A0K8WCS7 A0A0L0BUE7 Q174K2 A0A1B0FAF2 A0A182GWU5 A0A1A9Z1P9 A0A1A9X8T9 A0A1B0C3B7 A0A084WIY5 A0A1A9WTD5 A0A1I8N819 A0A336KVU8 A0A1A9VRK7 A0A182IRZ1 A0A2M3ZJ73 A0A2M3ZJ63 A0A1Q3F9X3 A0A2M4BUC4 A0A2M4BUB4 W5JGV7 A0A182R170 A0A2M4ARF4 B0WBV2 A0A1I8PSF3 A0A1I8PSK3 A0A182JZV1 A0A182X066 A0A182KT88 A0A182HPL2 A0A1I8PSI1 A0A182P1F7 A0A182YC88 A0A182M742 A0A182FTY4 A0A182N3R5 B4L2H9 B4M7V9 A0A0L7QQR6 Q7PZV2 A0A154PH42 T1PHY5 B4Q2U6 A0A2A3EE76 A0A088ASJ7 B3NTA7 U5EJQ4 B4I6N7 A0A0M4EUQ8 A0A1W4V0R5 H1UUC4 Q9VWY6 D3TQ88 B3MXU6 E2ACF0 A0A182RAI0 A0A195BBZ0 A0A158NMU2 A0A0M9ABS9 A0A195EXV9 B4JXF0 Q29H69 A0A151WUP8 A0A0J7NRY3 B4H082 A0A151JBE8 F4WTB0 A0A3L8DKT6 A0A026WM59 B4NQ20 A0A195C7L0 A0A2P8YHZ8 K7IMW3 E2BTN5 A0A182TEM4 A0A182VFQ5 A0A2J7PX97 A0A1J1HHU9 E0VGZ1 A0A067QYI5 A0A1W4XLI9 B0WBV1 A0A069DQF3 A0A232FF83 A0A023F9C6 A0A224XVQ6
I4DRV0 A0A3S2NBE9 S4PSZ0 H9JG50 A0A1L8DI39 A0A034WFA8 W8C8M9 A0A0K8WDZ2 A0A0K8WCS7 A0A0L0BUE7 Q174K2 A0A1B0FAF2 A0A182GWU5 A0A1A9Z1P9 A0A1A9X8T9 A0A1B0C3B7 A0A084WIY5 A0A1A9WTD5 A0A1I8N819 A0A336KVU8 A0A1A9VRK7 A0A182IRZ1 A0A2M3ZJ73 A0A2M3ZJ63 A0A1Q3F9X3 A0A2M4BUC4 A0A2M4BUB4 W5JGV7 A0A182R170 A0A2M4ARF4 B0WBV2 A0A1I8PSF3 A0A1I8PSK3 A0A182JZV1 A0A182X066 A0A182KT88 A0A182HPL2 A0A1I8PSI1 A0A182P1F7 A0A182YC88 A0A182M742 A0A182FTY4 A0A182N3R5 B4L2H9 B4M7V9 A0A0L7QQR6 Q7PZV2 A0A154PH42 T1PHY5 B4Q2U6 A0A2A3EE76 A0A088ASJ7 B3NTA7 U5EJQ4 B4I6N7 A0A0M4EUQ8 A0A1W4V0R5 H1UUC4 Q9VWY6 D3TQ88 B3MXU6 E2ACF0 A0A182RAI0 A0A195BBZ0 A0A158NMU2 A0A0M9ABS9 A0A195EXV9 B4JXF0 Q29H69 A0A151WUP8 A0A0J7NRY3 B4H082 A0A151JBE8 F4WTB0 A0A3L8DKT6 A0A026WM59 B4NQ20 A0A195C7L0 A0A2P8YHZ8 K7IMW3 E2BTN5 A0A182TEM4 A0A182VFQ5 A0A2J7PX97 A0A1J1HHU9 E0VGZ1 A0A067QYI5 A0A1W4XLI9 B0WBV1 A0A069DQF3 A0A232FF83 A0A023F9C6 A0A224XVQ6
PDB
5FUR
E-value=1.64196e-48,
Score=484
Ontologies
GO
PANTHER
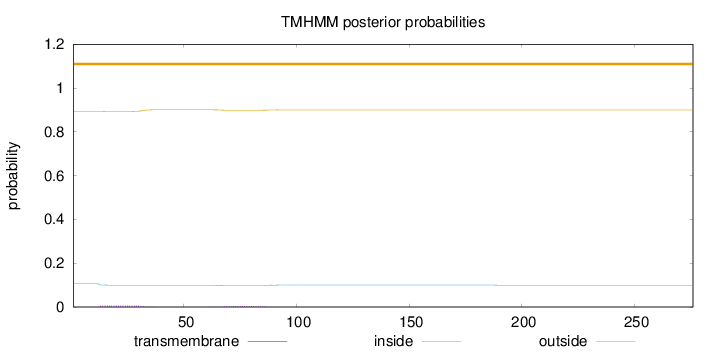
Topology
Subcellular location
Nucleus
Length:
276
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.20904
Exp number, first 60 AAs:
0.13394
Total prob of N-in:
0.10625
outside
1 - 276
Population Genetic Test Statistics
Pi
2.642004
Theta
31.690037
Tajima's D
-0.418974
CLR
1213.279126
CSRT
0.262386880655967
Interpretation
Uncertain
