Gene
KWMTBOMO10934
Annotation
PREDICTED:_mitochondrial_tRNA-specific_2-thiouridylase_1_[Bombyx_mori]
Full name
Mitochondrial tRNA-specific 2-thiouridylase 1
Location in the cell
Cytoplasmic Reliability : 2.418
Sequence
CDS
ATGTTTCCTCTTGCCAATTTATTAAAAACTGAAGTGAGAGAAATTGCTAAAAAGGAAGGTCTTCTAAATGTAGCGAATAAAAAAGATAGCACAGGAATATGCTTCATAGGTAAAAAAAAGTTTCAAGATTTTATTGCAGAATACATTGATACAAAAAAAGGACAATTCATAGATATAGATACTGGAAGGATTGTTGGTGTACACGAAGGTTTGCACAAGTGGACAGTAGGTCAAAGATGTTGCCTGCCCAATTGTAGAGATGCATATTTTGTATTGAAGAAAGACTTGGCAACGAATAACATTTATGTGGTTGCAGGAACAAAACATTCAGCATTGTGGAACAATATTTGCATCACTATGCCACCTCACTGGATTAGGAGAGAACCTCTTGAATTAACAGAAAATGGGGTGCTTAATTGTTACTTTAGGTTCCAGCATACAAAACCTTTAGCTCCATGCAAAACTGTGAAAAATTCCGAGGGACTTACTATCATCTTAAAAAATAATTTAAGAGCAATTACTGAAGGACAATATGCAGTACTCTATAAAGATGGAGAATGTCTTGGTAGTGCCAAAATGAAAAACATTTGTCACAATTTTGTTTTTTAA
Protein
MFPLANLLKTEVREIAKKEGLLNVANKKDSTGICFIGKKKFQDFIAEYIDTKKGQFIDIDTGRIVGVHEGLHKWTVGQRCCLPNCRDAYFVLKKDLATNNIYVVAGTKHSALWNNICITMPPHWIRREPLELTENGVLNCYFRFQHTKPLAPCKTVKNSEGLTIILKNNLRAITEGQYAVLYKDGECLGSAKMKNICHNFVF
Summary
Description
Catalyzes the 2-thiolation of uridine at the wobble position (U34) of mitochondrial tRNA(Lys), tRNA(Glu) and tRNA(Gln). Required for the formation of 5-taurinomethyl-2-thiouridine (tm5s2U) of mitochondrial tRNA(Lys), tRNA(Glu), and tRNA(Gln) at the wobble position. ATP is required to activate the C2 atom of the wobble base.
Catalytic Activity
5-taurinomethyluridine(34) in tRNA + AH2 + ATP + S-sulfanyl-L-cysteinyl-[protein] = 5-taurinomethyl-2-thiouridine(34) in tRNA + A + AMP + diphosphate + H(+) + L-cysteinyl-[protein]
Miscellaneous
During the reaction, ATP is used to activate the C2 atom of U34 by adenylation. After this, the persulfide sulfur on the catalytic cysteine is transferred to the C2 atom of the wobble base (U34) of mitochondrial tRNA(Lys), tRNA(Glu) and tRNA(Gln). The reaction probably involves hydrogen sulfide that is generated from the persulfide intermediate and that acts as nucleophile towards the activated C2 atom on U34. Subsequently, a transient disulfide bond is formed between the two active site cysteine residues (By similarity).
Similarity
Belongs to the MnmA/TRMU family.
Keywords
ATP-binding
Complete proteome
Disulfide bond
Mitochondrion
Nucleotide-binding
Reference proteome
RNA-binding
Transferase
tRNA processing
tRNA-binding
Feature
chain Mitochondrial tRNA-specific 2-thiouridylase 1
Uniprot
A0A2W1BRV0
A0A2H1VJ37
A0A2A4K4S1
A0A3S2NRE3
A0A194PGN6
A0A194RQD2
+ More
D6WSD8 A0A0N8CG99 A0A0P5T4C4 A0A0P6JLT6 A0A0P6B286 A0A0P5JAY7 A0A151XHF7 A0A154PQR8 A0A0L0CFM8 A0A195DJZ1 A0A2P8YGY5 A0A1I8P8S0 A0A1I8P8N8 A0A195C187 A0A195C2E6 A0A034WMB2 A0A151I1A0 N6TZ07 B4PXK2 A0A0K8VZ24 E9HGG5 A0A026VY94 Q9W5B6 A0A1Y1MTA8 A0A3L8D5W4 Q9U1L1 B4R2L6 A0A1J1IHL9 B3P9D2 A0A1I8MS83 B4I993 Q16EN5 Q170F2 K7J2F8 A0A232F4W9 A0A034WJT0 A0A2J7QWJ3 B0WBE3 A0A3R7PHY2 W8BN71 A0A1B6EMU6 A0A0K8TY76 A0A182GR64 W8C305 A0A2A3EAR4 A0A2M4CJ95 E2AFC4 A0A2T7NUH3 A0A1A9YMF3 E2C8U0 A0A0L7R3U6 A0A0V0GCD0 A0A1B3PDJ2 A0A1B6M3Q8 A0A0P4WJK1 A0A1B0D5K8 A0A182FV46 A0A1B6CLL6 A0A2M4CIT6 A0A1B0BDJ6 B3MYE9 A0A1B6DM22 A0A1B6C1U3 A0A3B0KYD4 A0A088AQD9 B4GTW3 A0A336KEP6 A0A224XLT6 B4L3S8 B5DNY9 A0A182TGC5 A0A0C9RVE2 Q7PZK6 A0A1S4H7Z4 A0A182HHP2 E9IA08 A0A182KAV0 A0A182L5A7 A0A0M5JA76 A0A182VK43 A0A1B0G8P8 A0A182XJN7 A0A1A9WVN2 A0A182QHX9 A0A182WE34 A0A2R7X028 B4JKU9 A0A182Y9K8 A0A1A9ZKS9 A0A182SUK1 B4N1Q7 A0A182N2Q0 A0A1W4VS81 A0A1B6C755
D6WSD8 A0A0N8CG99 A0A0P5T4C4 A0A0P6JLT6 A0A0P6B286 A0A0P5JAY7 A0A151XHF7 A0A154PQR8 A0A0L0CFM8 A0A195DJZ1 A0A2P8YGY5 A0A1I8P8S0 A0A1I8P8N8 A0A195C187 A0A195C2E6 A0A034WMB2 A0A151I1A0 N6TZ07 B4PXK2 A0A0K8VZ24 E9HGG5 A0A026VY94 Q9W5B6 A0A1Y1MTA8 A0A3L8D5W4 Q9U1L1 B4R2L6 A0A1J1IHL9 B3P9D2 A0A1I8MS83 B4I993 Q16EN5 Q170F2 K7J2F8 A0A232F4W9 A0A034WJT0 A0A2J7QWJ3 B0WBE3 A0A3R7PHY2 W8BN71 A0A1B6EMU6 A0A0K8TY76 A0A182GR64 W8C305 A0A2A3EAR4 A0A2M4CJ95 E2AFC4 A0A2T7NUH3 A0A1A9YMF3 E2C8U0 A0A0L7R3U6 A0A0V0GCD0 A0A1B3PDJ2 A0A1B6M3Q8 A0A0P4WJK1 A0A1B0D5K8 A0A182FV46 A0A1B6CLL6 A0A2M4CIT6 A0A1B0BDJ6 B3MYE9 A0A1B6DM22 A0A1B6C1U3 A0A3B0KYD4 A0A088AQD9 B4GTW3 A0A336KEP6 A0A224XLT6 B4L3S8 B5DNY9 A0A182TGC5 A0A0C9RVE2 Q7PZK6 A0A1S4H7Z4 A0A182HHP2 E9IA08 A0A182KAV0 A0A182L5A7 A0A0M5JA76 A0A182VK43 A0A1B0G8P8 A0A182XJN7 A0A1A9WVN2 A0A182QHX9 A0A182WE34 A0A2R7X028 B4JKU9 A0A182Y9K8 A0A1A9ZKS9 A0A182SUK1 B4N1Q7 A0A182N2Q0 A0A1W4VS81 A0A1B6C755
EC Number
2.8.1.14
Pubmed
EMBL
KZ149920
PZC77768.1
ODYU01002848
SOQ40838.1
NWSH01000165
PCG78908.1
+ More
RSAL01000123 RVE46635.1 KQ459604 KPI92556.1 KQ459765 KPJ20073.1 KQ971352 EFA07642.2 GDIP01134655 JAL69059.1 GDIP01131221 LRGB01001005 JAL72493.1 KZS14165.1 GDIQ01014631 JAN80106.1 GDIP01022388 JAM81327.1 GDIQ01227664 JAK24061.1 KQ982130 KYQ59844.1 KQ435036 KZC14077.1 JRES01000455 KNC31046.1 KQ980800 KYN12809.1 PYGN01000601 PSN43522.1 KQ978379 KYM94385.1 KYM94386.1 GAKP01004024 JAC54928.1 KQ976599 KYM79517.1 APGK01045809 APGK01045810 KB741039 ENN74530.1 CM000162 EDX00855.1 GDHF01014660 GDHF01008215 JAI37654.1 JAI44099.1 GL732641 EFX69171.1 KK107597 EZA48605.1 AE014298 AY051773 GEZM01024758 JAV87725.1 QOIP01000013 RLU15403.1 AL109630 CAB65878.1 CM000366 EDX16816.1 CVRI01000052 CRK99759.1 CH954183 EDV45428.1 CH480825 EDW43774.1 CH478641 EAT32694.1 EAT32696.1 EJY58120.1 CH477475 EAT40320.1 NNAY01000959 OXU25725.1 GAKP01004023 JAC54929.1 NEVH01009430 PNF32963.1 DS231879 EDS42382.1 QCYY01002846 ROT67123.1 GAMC01006303 JAC00253.1 GECZ01030538 JAS39231.1 GDHF01032880 JAI19434.1 JXUM01081860 KQ563273 KXJ74144.1 GAMC01006299 JAC00257.1 KZ288317 PBC28299.1 GGFL01001053 MBW65231.1 GL439108 EFN67817.1 PZQS01000009 PVD24838.1 GL453694 EFN75657.1 KQ414661 KOC65560.1 GECL01000630 JAP05494.1 KU659999 AOG17797.1 GEBQ01009447 JAT30530.1 GDRN01067963 GDRN01067961 GDRN01067960 GDRN01067959 JAI64279.1 AJVK01025409 GEDC01026878 GEDC01023117 JAS10420.1 JAS14181.1 GGFL01001078 MBW65256.1 JXJN01012503 CH902632 EDV32643.1 GEDC01010574 JAS26724.1 GEDC01029830 GEDC01015228 JAS07468.1 JAS22070.1 OUUW01000018 SPP89108.1 CH479190 EDW25983.1 UFQS01000328 UFQT01000328 SSX02921.1 SSX23288.1 GFTR01005638 JAW10788.1 CH933810 EDW07206.2 CH379065 EDY73241.1 GBYB01012870 JAG82637.1 AAAB01008986 EAA00244.3 APCN01002431 GL761885 EFZ22595.1 CP012528 ALC48161.1 CCAG010000299 AXCN02001081 KK856153 PTY25041.1 CH916370 EDW00202.1 CH963925 EDW78296.1 GEDC01027975 JAS09323.1
RSAL01000123 RVE46635.1 KQ459604 KPI92556.1 KQ459765 KPJ20073.1 KQ971352 EFA07642.2 GDIP01134655 JAL69059.1 GDIP01131221 LRGB01001005 JAL72493.1 KZS14165.1 GDIQ01014631 JAN80106.1 GDIP01022388 JAM81327.1 GDIQ01227664 JAK24061.1 KQ982130 KYQ59844.1 KQ435036 KZC14077.1 JRES01000455 KNC31046.1 KQ980800 KYN12809.1 PYGN01000601 PSN43522.1 KQ978379 KYM94385.1 KYM94386.1 GAKP01004024 JAC54928.1 KQ976599 KYM79517.1 APGK01045809 APGK01045810 KB741039 ENN74530.1 CM000162 EDX00855.1 GDHF01014660 GDHF01008215 JAI37654.1 JAI44099.1 GL732641 EFX69171.1 KK107597 EZA48605.1 AE014298 AY051773 GEZM01024758 JAV87725.1 QOIP01000013 RLU15403.1 AL109630 CAB65878.1 CM000366 EDX16816.1 CVRI01000052 CRK99759.1 CH954183 EDV45428.1 CH480825 EDW43774.1 CH478641 EAT32694.1 EAT32696.1 EJY58120.1 CH477475 EAT40320.1 NNAY01000959 OXU25725.1 GAKP01004023 JAC54929.1 NEVH01009430 PNF32963.1 DS231879 EDS42382.1 QCYY01002846 ROT67123.1 GAMC01006303 JAC00253.1 GECZ01030538 JAS39231.1 GDHF01032880 JAI19434.1 JXUM01081860 KQ563273 KXJ74144.1 GAMC01006299 JAC00257.1 KZ288317 PBC28299.1 GGFL01001053 MBW65231.1 GL439108 EFN67817.1 PZQS01000009 PVD24838.1 GL453694 EFN75657.1 KQ414661 KOC65560.1 GECL01000630 JAP05494.1 KU659999 AOG17797.1 GEBQ01009447 JAT30530.1 GDRN01067963 GDRN01067961 GDRN01067960 GDRN01067959 JAI64279.1 AJVK01025409 GEDC01026878 GEDC01023117 JAS10420.1 JAS14181.1 GGFL01001078 MBW65256.1 JXJN01012503 CH902632 EDV32643.1 GEDC01010574 JAS26724.1 GEDC01029830 GEDC01015228 JAS07468.1 JAS22070.1 OUUW01000018 SPP89108.1 CH479190 EDW25983.1 UFQS01000328 UFQT01000328 SSX02921.1 SSX23288.1 GFTR01005638 JAW10788.1 CH933810 EDW07206.2 CH379065 EDY73241.1 GBYB01012870 JAG82637.1 AAAB01008986 EAA00244.3 APCN01002431 GL761885 EFZ22595.1 CP012528 ALC48161.1 CCAG010000299 AXCN02001081 KK856153 PTY25041.1 CH916370 EDW00202.1 CH963925 EDW78296.1 GEDC01027975 JAS09323.1
Proteomes
UP000218220
UP000283053
UP000053268
UP000053240
UP000007266
UP000076858
+ More
UP000075809 UP000076502 UP000037069 UP000078492 UP000245037 UP000095300 UP000078542 UP000078540 UP000019118 UP000002282 UP000000305 UP000053097 UP000000803 UP000279307 UP000000304 UP000183832 UP000008711 UP000095301 UP000001292 UP000008820 UP000002358 UP000215335 UP000235965 UP000002320 UP000283509 UP000069940 UP000249989 UP000242457 UP000000311 UP000245119 UP000092443 UP000008237 UP000053825 UP000092462 UP000069272 UP000092460 UP000007801 UP000268350 UP000005203 UP000008744 UP000009192 UP000001819 UP000075902 UP000007062 UP000075840 UP000075881 UP000075882 UP000092553 UP000075903 UP000092444 UP000076407 UP000091820 UP000075886 UP000075920 UP000001070 UP000076408 UP000092445 UP000075901 UP000007798 UP000075884 UP000192221
UP000075809 UP000076502 UP000037069 UP000078492 UP000245037 UP000095300 UP000078542 UP000078540 UP000019118 UP000002282 UP000000305 UP000053097 UP000000803 UP000279307 UP000000304 UP000183832 UP000008711 UP000095301 UP000001292 UP000008820 UP000002358 UP000215335 UP000235965 UP000002320 UP000283509 UP000069940 UP000249989 UP000242457 UP000000311 UP000245119 UP000092443 UP000008237 UP000053825 UP000092462 UP000069272 UP000092460 UP000007801 UP000268350 UP000005203 UP000008744 UP000009192 UP000001819 UP000075902 UP000007062 UP000075840 UP000075881 UP000075882 UP000092553 UP000075903 UP000092444 UP000076407 UP000091820 UP000075886 UP000075920 UP000001070 UP000076408 UP000092445 UP000075901 UP000007798 UP000075884 UP000192221
Pfam
PF00085 Thioredoxin
Interpro
SUPFAM
SSF52833
SSF52833
Gene 3D
ProteinModelPortal
A0A2W1BRV0
A0A2H1VJ37
A0A2A4K4S1
A0A3S2NRE3
A0A194PGN6
A0A194RQD2
+ More
D6WSD8 A0A0N8CG99 A0A0P5T4C4 A0A0P6JLT6 A0A0P6B286 A0A0P5JAY7 A0A151XHF7 A0A154PQR8 A0A0L0CFM8 A0A195DJZ1 A0A2P8YGY5 A0A1I8P8S0 A0A1I8P8N8 A0A195C187 A0A195C2E6 A0A034WMB2 A0A151I1A0 N6TZ07 B4PXK2 A0A0K8VZ24 E9HGG5 A0A026VY94 Q9W5B6 A0A1Y1MTA8 A0A3L8D5W4 Q9U1L1 B4R2L6 A0A1J1IHL9 B3P9D2 A0A1I8MS83 B4I993 Q16EN5 Q170F2 K7J2F8 A0A232F4W9 A0A034WJT0 A0A2J7QWJ3 B0WBE3 A0A3R7PHY2 W8BN71 A0A1B6EMU6 A0A0K8TY76 A0A182GR64 W8C305 A0A2A3EAR4 A0A2M4CJ95 E2AFC4 A0A2T7NUH3 A0A1A9YMF3 E2C8U0 A0A0L7R3U6 A0A0V0GCD0 A0A1B3PDJ2 A0A1B6M3Q8 A0A0P4WJK1 A0A1B0D5K8 A0A182FV46 A0A1B6CLL6 A0A2M4CIT6 A0A1B0BDJ6 B3MYE9 A0A1B6DM22 A0A1B6C1U3 A0A3B0KYD4 A0A088AQD9 B4GTW3 A0A336KEP6 A0A224XLT6 B4L3S8 B5DNY9 A0A182TGC5 A0A0C9RVE2 Q7PZK6 A0A1S4H7Z4 A0A182HHP2 E9IA08 A0A182KAV0 A0A182L5A7 A0A0M5JA76 A0A182VK43 A0A1B0G8P8 A0A182XJN7 A0A1A9WVN2 A0A182QHX9 A0A182WE34 A0A2R7X028 B4JKU9 A0A182Y9K8 A0A1A9ZKS9 A0A182SUK1 B4N1Q7 A0A182N2Q0 A0A1W4VS81 A0A1B6C755
D6WSD8 A0A0N8CG99 A0A0P5T4C4 A0A0P6JLT6 A0A0P6B286 A0A0P5JAY7 A0A151XHF7 A0A154PQR8 A0A0L0CFM8 A0A195DJZ1 A0A2P8YGY5 A0A1I8P8S0 A0A1I8P8N8 A0A195C187 A0A195C2E6 A0A034WMB2 A0A151I1A0 N6TZ07 B4PXK2 A0A0K8VZ24 E9HGG5 A0A026VY94 Q9W5B6 A0A1Y1MTA8 A0A3L8D5W4 Q9U1L1 B4R2L6 A0A1J1IHL9 B3P9D2 A0A1I8MS83 B4I993 Q16EN5 Q170F2 K7J2F8 A0A232F4W9 A0A034WJT0 A0A2J7QWJ3 B0WBE3 A0A3R7PHY2 W8BN71 A0A1B6EMU6 A0A0K8TY76 A0A182GR64 W8C305 A0A2A3EAR4 A0A2M4CJ95 E2AFC4 A0A2T7NUH3 A0A1A9YMF3 E2C8U0 A0A0L7R3U6 A0A0V0GCD0 A0A1B3PDJ2 A0A1B6M3Q8 A0A0P4WJK1 A0A1B0D5K8 A0A182FV46 A0A1B6CLL6 A0A2M4CIT6 A0A1B0BDJ6 B3MYE9 A0A1B6DM22 A0A1B6C1U3 A0A3B0KYD4 A0A088AQD9 B4GTW3 A0A336KEP6 A0A224XLT6 B4L3S8 B5DNY9 A0A182TGC5 A0A0C9RVE2 Q7PZK6 A0A1S4H7Z4 A0A182HHP2 E9IA08 A0A182KAV0 A0A182L5A7 A0A0M5JA76 A0A182VK43 A0A1B0G8P8 A0A182XJN7 A0A1A9WVN2 A0A182QHX9 A0A182WE34 A0A2R7X028 B4JKU9 A0A182Y9K8 A0A1A9ZKS9 A0A182SUK1 B4N1Q7 A0A182N2Q0 A0A1W4VS81 A0A1B6C755
PDB
2DEU
E-value=2.31008e-27,
Score=300
Ontologies
GO
Topology
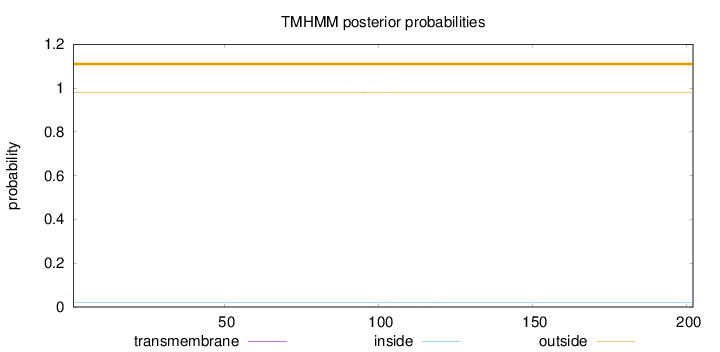
Subcellular location
Mitochondrion
Length:
202
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00617
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01975
outside
1 - 202
Population Genetic Test Statistics
Pi
157.366853
Theta
185.060735
Tajima's D
-0.648471
CLR
132.840239
CSRT
0.210939453027349
Interpretation
Uncertain
