Gene
KWMTBOMO10933
Annotation
PREDICTED:_ribonuclease_3_[Bombyx_mori]
Full name
XK-related protein
Location in the cell
Nuclear Reliability : 4.089
Sequence
CDS
ATGTCTATTCCTCCAGTTGATTACAGTGTTCCACCTCCGCCAATTCCGAGGCTTCCAGTTAATATTCCGTTACCACCCCACCCACCCCCGCCTATCTTTCCGAGAGCAATGCCTTTATATACCAGACCGCCACCAGTACTTCCGATGCTATCAGTACCGCCAGTACCATACCCTCATTCACGCATGCCTCCAGCTCCTGTTCCACAGTTACCTGTACCCAACTTTATGCCCACCATTGATATACATTCTGTCCCTCCACCACATCCATCCCCTCACATAGGACATTCTCCACATGGATATTCTGATAGGAGCAGGAGGGAGAGGGATAGAGATAGAGAACAGAGGAATCCGACAAGTCATAGTCCTAGATATTGCTCTAGAAGTCCACCAAGATACAGTTCAAAATTAAACAGAAGTCCTTCTCGATATAGCTATAGAGAAAGAGAGAAAGAAAGATATTACATGGAGAGAGAGAAAAAGTATTATGAAGGAAGACGAGAATATGGTAGAAGAAAATATGAATATGATGTTCGCCGCAGTGAATCTCAATACAAAGATTATAGAAGCCACAGTAGATGTGAAGGTAGGGATAGATCACCAAGCAGAAGGGCGCATTCTCCTAGTAGGGAGTCCAATAGAGAAAGTCTGACGAGAGAAATGTCACTTCACCGACAAGAATCACTTTTAGGCAAACCAATGACAGATCGTGAAAAGATTTTGGAAGAATATAGGAAAAACTACTGTAGAACTTCTGAAGATTTTATACAGAAAATGGAACAGTGGTCGAAAGAACATCTTTCTGAAGGAGAAGAGTCGTCAATGTGGTATAGGAGTTCACCAGCTGAGCTCTATTACAAAGAAGTCCAAAATGGTGAGGGAGTCGAACAAACTTTAAAACTGCAGAAACTTTGTGAAGTATTCATGAAGAAACTCATTGAGAGGGGACGTAATGCAAGGCCTGAGGTTGATGAGCTCCCACCCTTAAAGCCACCTAAATCGAAAATGTGCAAACATAAACTTGATGAAAAAAGCTCATCGTCGTCATCATCGTCATCATCATCAGAAAGTGACGGATGTTTAGATGAAGAGGGCTTGCAGGGCTATAGTGATAGGATAATGCTAGAGCTACAGCGGAAACAGAGTCATCCCAGGAGATTGCACCCTGAAATGTGGTTTAACAACACCGGCGAAATGAACGGCGGTCCGTTGTGTCGGTGTAGCGCCCGAGCCCGTCGTCACGGCATGCGGCACGGCGTCTACGCCGGCGAGGAATCATTCCCGAAATGTATTCCTGAACGAAGCAATATCGACAAACTGTATCATTATCGTATAACCGTGTCGCCTCCTACAAACTTTTTAATCAAAGCACCAACGATCATAGCTCACGACGAACACGAATTTCTATTTTCTGGCTTCTCATTGTTCTCGCACTACAAACTTGCCAAGTTGCCCATGTGCAAAGTCATAAGGTTCAATATTGAATACACAATTCTATATGTGGAAGAACCGGCACCAAGAAATTTTACCATACAAGAATTAAGTCTATTCGAGGAATTCCTATTTACGGAGTTACTAGAGTTGATTGATTTGGACCTGGGAAAAGCCACGGATACAACATGTTCACAATTCCATTTTATGCCTCGATTTGTAAGATTTTTACCTGAAGGAGGATGTGAGGTACTCTCAATGTGTGAAGTTATAAAGTATTTGATTGACGAAAGTGGTTTACTGATACCGATGGAAAAGTTGAGTGACGTACATGAAATGGATCACTACACCTGGCAAAAGTTTGTTGATAAAATCAAAGGTATGATAGTGACTTACCCCGGTAAGAAGCCGTGCTCGGTGCGCGTGGACCAGCTCGACCGAAGTCCGCCGACGGGCTCCGAAGCCGTTAAGAACCCTAAAGCATTGAAGAAATACTTCCCTGAAATCGTACACTTTGGTATACGTCCGCCGCAGCTCAGTTACGCTGGCAACCCCGAGTATCAAAAAGCATGGCGTTATTACGTTAAATACCGCCATTTAATAGCGAATATGGCGAAGACTTCTTATAAGGAGAGACAGAAATTAGCCGCTAAAGAGGCTAAGCTGCAAGAGATGCGCACTCAGTCTAAAATGAAAAGGGACGTGACTGTCGCTATATCGTCGGAAGGGTTCCACACTACCGGACTCATGTGTGACGTCGTGCAGCACGCGATGTTAATACCGGTGTTGGTGCGTCACCTCCGGTTCCACAAGTCGCTCGAAAAACTCGAGAAGAAAATCGGTTATGTTTTTAAGCAACGAGCATTGCTCCAGACTGCATTAACTCATCCATCGTATAGAGAAAACTTTGGTACCAACCCTGACCACGCCAGGAACAGTCTCACTAACTGCGGTATACGACAACCGGAGTACGGGGACCGAAGAATACATTATACAAGAAAGAAGGGCATAGTAACACTCATCAAAATAATGTCCAGTTTCGGGAAAATCAGTGAGACAGAATCAGAGATTAAGCACAACGAACGTTTGGAATTTTTGGGAGATGCTGTAGTGGAATTCTTGTCTTCGATTCATCTTTTTAAGATGTTTCCTGGGCTGGCAGAAGGAGGACTCGCTACTTACAGAGCTGCCATTGTACAGAATCAACATTTAGCGCAACTTGCTAAGAATATAAACCTTGAACAATTCATGTTGTACGCCCACGGTTCGGACCTGTGTCGCGACGTAGTGATGCGACACGCGATGGCGAACTGCTTCGAAGCTCTCATGGGCGCATTGTTTCTCGACGCTGGACTTGAGGTAGCGGACCGAGTCTTCTCTTTGGCATTGTGGCACAATGAATCGGAGCTGCTTGAAGTTTGGACGAAGGAAAGAGCACATCCATTACAAGAACAAGAACCTTTAGGCGATAGGAAATATATAAAAGACTTCGAATTCTTACAAAAATTGACCGACTTCGAAGAATCTATAGGGGTACAGTTCAAACACATACGTTTGTTGGCTCGTGCGTTCACGGATCGCTCAGTCGGCTTCACTCTCCTCACGCTCGGATCCAACCAGCGCTTAGAGTTCCTCGGCGACACGGTGCTCCAGCTGGTCGTGTCCGATCGACTGTACAGACACTTCCCTGATCATCACGAAGGCCATTTGTCGCTGTTACGGTCGTCTCTGGTCAACAACAGAACACAGTCTATGGTTTGCGACGATCTGAACATGTCTGCGTACGCCATATACAATAATCCTAAAGCGAAACCGACCACAAAGAAACATAAGGCTGATTTACTCGAGGCTTTCTTGGGAGCATTGTTTATTGATAAGAACCTGGAGTACTGTCAGGCGTTCTGCAACGCGTGTCTGTTTCCGCGCCTGCAGCAGTTCATCATGAACCAAGACTGGAACGATCCCAAGTCTAAATTACAGCAGTGCTGCCTCACGTTGCGGTCCATGGAGGGGGGCGAGCCGGACATACCAGTCTACAAAGTAATCCAGTGTCTGGGACCTACGAACACGCGAGTGTATACTGTGGGCGTGTATTTCCGCGGGAAGAGATTGGCGGCCGCACGCGGACACTCCATACAGGAGGCCGAGATGAACGCGGCCGAAGAAGCCTTGAATACTGCATACGGTTTATTCCCGCAATTAGATCACCAGAAACGTATAATAGCGAAGAGCGTACGTAAGAAGAAGGGGGATCGCGAGGATCCGTCCGAGAATAGTTCGCACGGAATGCACGACGACAACGTACCGAAAGCTTACAGGCTCGGCAGCAAACCTGACAGTTCTGATGAAAGCAGCGTCCACAGCGGATCCGAGGATCCCGAGGATCCCGAGTCGCCGGCCAAATCGGACGAGGACAGCGGCCTAGACAGCGACACCGAGTCGGAGCTGCCGGGCAAACTCAAAAACGTCCAAGTCAGCAGTCTGCTCAGCGACATGGAGAAAATAAAACAGGAACTCATCAAGAACAACGAGTATGAGAAACTCAAGGAGAAATGCAGACGGTCCGACTCAGATGAAGAATGA
Protein
MSIPPVDYSVPPPPIPRLPVNIPLPPHPPPPIFPRAMPLYTRPPPVLPMLSVPPVPYPHSRMPPAPVPQLPVPNFMPTIDIHSVPPPHPSPHIGHSPHGYSDRSRRERDRDREQRNPTSHSPRYCSRSPPRYSSKLNRSPSRYSYREREKERYYMEREKKYYEGRREYGRRKYEYDVRRSESQYKDYRSHSRCEGRDRSPSRRAHSPSRESNRESLTREMSLHRQESLLGKPMTDREKILEEYRKNYCRTSEDFIQKMEQWSKEHLSEGEESSMWYRSSPAELYYKEVQNGEGVEQTLKLQKLCEVFMKKLIERGRNARPEVDELPPLKPPKSKMCKHKLDEKSSSSSSSSSSSESDGCLDEEGLQGYSDRIMLELQRKQSHPRRLHPEMWFNNTGEMNGGPLCRCSARARRHGMRHGVYAGEESFPKCIPERSNIDKLYHYRITVSPPTNFLIKAPTIIAHDEHEFLFSGFSLFSHYKLAKLPMCKVIRFNIEYTILYVEEPAPRNFTIQELSLFEEFLFTELLELIDLDLGKATDTTCSQFHFMPRFVRFLPEGGCEVLSMCEVIKYLIDESGLLIPMEKLSDVHEMDHYTWQKFVDKIKGMIVTYPGKKPCSVRVDQLDRSPPTGSEAVKNPKALKKYFPEIVHFGIRPPQLSYAGNPEYQKAWRYYVKYRHLIANMAKTSYKERQKLAAKEAKLQEMRTQSKMKRDVTVAISSEGFHTTGLMCDVVQHAMLIPVLVRHLRFHKSLEKLEKKIGYVFKQRALLQTALTHPSYRENFGTNPDHARNSLTNCGIRQPEYGDRRIHYTRKKGIVTLIKIMSSFGKISETESEIKHNERLEFLGDAVVEFLSSIHLFKMFPGLAEGGLATYRAAIVQNQHLAQLAKNINLEQFMLYAHGSDLCRDVVMRHAMANCFEALMGALFLDAGLEVADRVFSLALWHNESELLEVWTKERAHPLQEQEPLGDRKYIKDFEFLQKLTDFEESIGVQFKHIRLLARAFTDRSVGFTLLTLGSNQRLEFLGDTVLQLVVSDRLYRHFPDHHEGHLSLLRSSLVNNRTQSMVCDDLNMSAYAIYNNPKAKPTTKKHKADLLEAFLGALFIDKNLEYCQAFCNACLFPRLQQFIMNQDWNDPKSKLQQCCLTLRSMEGGEPDIPVYKVIQCLGPTNTRVYTVGVYFRGKRLAAARGHSIQEAEMNAAEEALNTAYGLFPQLDHQKRIIAKSVRKKKGDREDPSENSSHGMHDDNVPKAYRLGSKPDSSDESSVHSGSEDPEDPESPAKSDEDSGLDSDTESELPGKLKNVQVSSLLSDMEKIKQELIKNNEYEKLKEKCRRSDSDEE
Summary
Similarity
Belongs to the XK family.
Uniprot
A0A2P1DM67
A0A2H1VJ32
A0A2W1BX45
A0A2A4K3R9
A0A212FC99
F4X4J6
+ More
A0A151JV17 A0A195BCU5 A0A158NIK6 A0A151XG45 A0A195CNP1 A0A232ELJ3 E2BFD6 K7J8Y7 A0A1W4XHW0 A0A0C9QGH1 A0A154P7A7 A0A3L8DNL8 A0A0J7KXI3 A0A0M8ZSV3 A0A026W231 A0A2P1DM47 A0A2J7R7F0 A0A2I6PIZ4 A0A0L7R5L2 A0A2A3EGM8 A0A088ACD3 A0A1Y1K9T9 A0A151J2X3 A0A310SU72 A0A067QJY1 A0A182T4I3 E9I8E2 A0A182Y052 A0A182U222 A0A182V1W0 A0A182WU62 A0A182J9R8 A0A1S4GX90 A0A182JTQ4 A0A182HUS4 Q5TQK8 A0A084VN71 A0A182L1K9 A0A182W763 A0A2R7VTY1 A0A182NUA6 A0A182PMM7 A0A182MAP4 A0A182QBA2 A0A182RTC0 A0A2P1DM88 A0A1Q3G295 A0A1B6C2Y5 A0A1S4J407 W5JRG0 A0A336LWE9 A0A336LTC6 A0A336MRR9 A0A1B0G5S3 A0A1A9US91 A0A0L0CPI3 A0A2M4A4G9 A0A182H177 A0A1A9ZAW6 A0A1A9XIV2 A0A1B0BUH8 Q16YA9 A0A1J1IPR8 A0A0K8V217 A0A182FJS9 A0A034W5R4 A0A023F4I7 A0A0A1XKY5 A0A224XDC0 A0A146L6Y6 W8CEJ8 B4KQ41 A0A1A9WEI4 A0A146KXR5 A0A0A9X7Y5 K7YZ60 T1I9Z1 B4J860 A0A0M3QVP3 B4NMS8 B4MF95 B3N9C5 A0A1I8NIB3 A0A3B0JPJ8 A0A3B0JJL1 A0A139WAR5 B4GD51 E0VZ53 Q28YB7 A0A1I8P8E5 B4P256 A0A1W4VZ00
A0A151JV17 A0A195BCU5 A0A158NIK6 A0A151XG45 A0A195CNP1 A0A232ELJ3 E2BFD6 K7J8Y7 A0A1W4XHW0 A0A0C9QGH1 A0A154P7A7 A0A3L8DNL8 A0A0J7KXI3 A0A0M8ZSV3 A0A026W231 A0A2P1DM47 A0A2J7R7F0 A0A2I6PIZ4 A0A0L7R5L2 A0A2A3EGM8 A0A088ACD3 A0A1Y1K9T9 A0A151J2X3 A0A310SU72 A0A067QJY1 A0A182T4I3 E9I8E2 A0A182Y052 A0A182U222 A0A182V1W0 A0A182WU62 A0A182J9R8 A0A1S4GX90 A0A182JTQ4 A0A182HUS4 Q5TQK8 A0A084VN71 A0A182L1K9 A0A182W763 A0A2R7VTY1 A0A182NUA6 A0A182PMM7 A0A182MAP4 A0A182QBA2 A0A182RTC0 A0A2P1DM88 A0A1Q3G295 A0A1B6C2Y5 A0A1S4J407 W5JRG0 A0A336LWE9 A0A336LTC6 A0A336MRR9 A0A1B0G5S3 A0A1A9US91 A0A0L0CPI3 A0A2M4A4G9 A0A182H177 A0A1A9ZAW6 A0A1A9XIV2 A0A1B0BUH8 Q16YA9 A0A1J1IPR8 A0A0K8V217 A0A182FJS9 A0A034W5R4 A0A023F4I7 A0A0A1XKY5 A0A224XDC0 A0A146L6Y6 W8CEJ8 B4KQ41 A0A1A9WEI4 A0A146KXR5 A0A0A9X7Y5 K7YZ60 T1I9Z1 B4J860 A0A0M3QVP3 B4NMS8 B4MF95 B3N9C5 A0A1I8NIB3 A0A3B0JPJ8 A0A3B0JJL1 A0A139WAR5 B4GD51 E0VZ53 Q28YB7 A0A1I8P8E5 B4P256 A0A1W4VZ00
Pubmed
28756777
22118469
21719571
21347285
28648823
20798317
+ More
20075255 30249741 24508170 29267401 28004739 24845553 21282665 25244985 12364791 24438588 20966253 20920257 23761445 26108605 26483478 17510324 25348373 25474469 25830018 26823975 24495485 17994087 25401762 23232437 25315136 18362917 19820115 20566863 15632085 17550304
20075255 30249741 24508170 29267401 28004739 24845553 21282665 25244985 12364791 24438588 20966253 20920257 23761445 26108605 26483478 17510324 25348373 25474469 25830018 26823975 24495485 17994087 25401762 23232437 25315136 18362917 19820115 20566863 15632085 17550304
EMBL
MG225429
AVK59440.1
ODYU01002848
SOQ40837.1
KZ149920
PZC77767.1
+ More
NWSH01000165 PCG78905.1 AGBW02009208 OWR51365.1 GL888656 EGI58626.1 KQ981733 KYN36551.1 KQ976513 KYM82391.1 ADTU01016899 ADTU01016900 ADTU01016901 KQ982179 KYQ59268.1 KQ977481 KYN02358.1 NNAY01003549 OXU19217.1 GL448009 EFN85596.1 GBYB01013653 JAG83420.1 KQ434831 KZC07809.1 QOIP01000006 RLU22040.1 LBMM01002320 KMQ94989.1 KQ435915 KOX68839.1 KK107503 EZA49641.1 MG225416 AVK59427.1 NEVH01006733 PNF36736.1 MG266020 AUM60047.1 KQ414654 KOC66051.1 KZ288256 PBC30634.1 GEZM01088399 GEZM01088398 JAV58223.1 KQ980336 KYN16488.1 KQ760132 OAD61835.1 KK853380 KDR08017.1 GL761538 EFZ23143.1 AAAB01008964 APCN01002476 EAL39656.3 ATLV01014722 KE524984 KFB39415.1 KK854051 PTY10331.1 AXCM01002726 AXCN02000770 MG225445 AVK59456.1 GFDL01001123 JAV33922.1 GEDC01029425 GEDC01009810 JAS07873.1 JAS27488.1 ADMH02000558 ETN65888.1 UFQT01000186 SSX21291.1 SSX21292.1 UFQS01002145 UFQT01002145 SSX13324.1 SSX32760.1 CCAG010015437 JRES01000105 KNC34082.1 GGFK01002385 MBW35706.1 JXUM01102762 KQ564750 KXJ71800.1 JXJN01020712 JXJN01025590 CH477519 EAT39617.1 CVRI01000056 CRL01714.1 GDHF01019679 JAI32635.1 GAKP01009325 JAC49627.1 GBBI01002595 JAC16117.1 GBXI01002283 JAD12009.1 GFTR01008648 JAW07778.1 GDHC01014435 JAQ04194.1 GAMC01000071 JAC06485.1 CH933808 EDW08143.1 GDHC01018693 JAP99935.1 GBHO01030493 GBHO01027858 GBHO01027855 GBHO01027854 JAG13111.1 JAG15746.1 JAG15749.1 JAG15750.1 JX679517 AFX89030.1 ACPB03008226 CH916367 EDW01197.1 CP012524 ALC42743.1 CH964282 EDW85667.1 CH940667 EDW57264.1 CH954177 EDV59612.1 OUUW01000001 SPP75549.1 SPP75550.1 KQ971380 KYB24989.1 CH479181 EDW32546.1 DS235849 EEB18659.1 CM000071 EAL26049.2 CM000157 EDW89257.1
NWSH01000165 PCG78905.1 AGBW02009208 OWR51365.1 GL888656 EGI58626.1 KQ981733 KYN36551.1 KQ976513 KYM82391.1 ADTU01016899 ADTU01016900 ADTU01016901 KQ982179 KYQ59268.1 KQ977481 KYN02358.1 NNAY01003549 OXU19217.1 GL448009 EFN85596.1 GBYB01013653 JAG83420.1 KQ434831 KZC07809.1 QOIP01000006 RLU22040.1 LBMM01002320 KMQ94989.1 KQ435915 KOX68839.1 KK107503 EZA49641.1 MG225416 AVK59427.1 NEVH01006733 PNF36736.1 MG266020 AUM60047.1 KQ414654 KOC66051.1 KZ288256 PBC30634.1 GEZM01088399 GEZM01088398 JAV58223.1 KQ980336 KYN16488.1 KQ760132 OAD61835.1 KK853380 KDR08017.1 GL761538 EFZ23143.1 AAAB01008964 APCN01002476 EAL39656.3 ATLV01014722 KE524984 KFB39415.1 KK854051 PTY10331.1 AXCM01002726 AXCN02000770 MG225445 AVK59456.1 GFDL01001123 JAV33922.1 GEDC01029425 GEDC01009810 JAS07873.1 JAS27488.1 ADMH02000558 ETN65888.1 UFQT01000186 SSX21291.1 SSX21292.1 UFQS01002145 UFQT01002145 SSX13324.1 SSX32760.1 CCAG010015437 JRES01000105 KNC34082.1 GGFK01002385 MBW35706.1 JXUM01102762 KQ564750 KXJ71800.1 JXJN01020712 JXJN01025590 CH477519 EAT39617.1 CVRI01000056 CRL01714.1 GDHF01019679 JAI32635.1 GAKP01009325 JAC49627.1 GBBI01002595 JAC16117.1 GBXI01002283 JAD12009.1 GFTR01008648 JAW07778.1 GDHC01014435 JAQ04194.1 GAMC01000071 JAC06485.1 CH933808 EDW08143.1 GDHC01018693 JAP99935.1 GBHO01030493 GBHO01027858 GBHO01027855 GBHO01027854 JAG13111.1 JAG15746.1 JAG15749.1 JAG15750.1 JX679517 AFX89030.1 ACPB03008226 CH916367 EDW01197.1 CP012524 ALC42743.1 CH964282 EDW85667.1 CH940667 EDW57264.1 CH954177 EDV59612.1 OUUW01000001 SPP75549.1 SPP75550.1 KQ971380 KYB24989.1 CH479181 EDW32546.1 DS235849 EEB18659.1 CM000071 EAL26049.2 CM000157 EDW89257.1
Proteomes
UP000218220
UP000007151
UP000007755
UP000078541
UP000078540
UP000005205
+ More
UP000075809 UP000078542 UP000215335 UP000008237 UP000002358 UP000192223 UP000076502 UP000279307 UP000036403 UP000053105 UP000053097 UP000235965 UP000053825 UP000242457 UP000005203 UP000078492 UP000027135 UP000075901 UP000076408 UP000075902 UP000075903 UP000076407 UP000075880 UP000075881 UP000075840 UP000007062 UP000030765 UP000075882 UP000075920 UP000075884 UP000075885 UP000075883 UP000075886 UP000075900 UP000000673 UP000092444 UP000078200 UP000037069 UP000069940 UP000249989 UP000092445 UP000092443 UP000092460 UP000008820 UP000183832 UP000069272 UP000009192 UP000091820 UP000015103 UP000001070 UP000092553 UP000007798 UP000008792 UP000008711 UP000095301 UP000268350 UP000007266 UP000008744 UP000009046 UP000001819 UP000095300 UP000002282 UP000192221
UP000075809 UP000078542 UP000215335 UP000008237 UP000002358 UP000192223 UP000076502 UP000279307 UP000036403 UP000053105 UP000053097 UP000235965 UP000053825 UP000242457 UP000005203 UP000078492 UP000027135 UP000075901 UP000076408 UP000075902 UP000075903 UP000076407 UP000075880 UP000075881 UP000075840 UP000007062 UP000030765 UP000075882 UP000075920 UP000075884 UP000075885 UP000075883 UP000075886 UP000075900 UP000000673 UP000092444 UP000078200 UP000037069 UP000069940 UP000249989 UP000092445 UP000092443 UP000092460 UP000008820 UP000183832 UP000069272 UP000009192 UP000091820 UP000015103 UP000001070 UP000092553 UP000007798 UP000008792 UP000008711 UP000095301 UP000268350 UP000007266 UP000008744 UP000009046 UP000001819 UP000095300 UP000002282 UP000192221
Pfam
Interpro
IPR000999
RNase_III_dom
+ More
IPR036389 RNase_III_sf
IPR014720 dsRBD_dom
IPR011907 RNase_III
IPR005225 Small_GTP-bd_dom
IPR001806 Small_GTPase
IPR027417 P-loop_NTPase
IPR012337 RNaseH-like_sf
IPR037056 RNase_H1_N_sf
IPR036397 RNaseH_sf
IPR002156 RNaseH_domain
IPR011320 RNase_H1_N
IPR009027 Ribosomal_L9/RNase_H1_N
IPR023674 Ribosomal_L1-like
IPR023673 Ribosomal_L1_CS
IPR016095 Ribosomal_L1_3-a/b-sand
IPR028364 Ribosomal_L1/biogenesis
IPR018629 XK-rel
IPR000372 LRRNT
IPR001611 Leu-rich_rpt
IPR020846 MFS_dom
IPR032675 LRR_dom_sf
IPR036389 RNase_III_sf
IPR014720 dsRBD_dom
IPR011907 RNase_III
IPR005225 Small_GTP-bd_dom
IPR001806 Small_GTPase
IPR027417 P-loop_NTPase
IPR012337 RNaseH-like_sf
IPR037056 RNase_H1_N_sf
IPR036397 RNaseH_sf
IPR002156 RNaseH_domain
IPR011320 RNase_H1_N
IPR009027 Ribosomal_L9/RNase_H1_N
IPR023674 Ribosomal_L1-like
IPR023673 Ribosomal_L1_CS
IPR016095 Ribosomal_L1_3-a/b-sand
IPR028364 Ribosomal_L1/biogenesis
IPR018629 XK-rel
IPR000372 LRRNT
IPR001611 Leu-rich_rpt
IPR020846 MFS_dom
IPR032675 LRR_dom_sf
SUPFAM
ProteinModelPortal
A0A2P1DM67
A0A2H1VJ32
A0A2W1BX45
A0A2A4K3R9
A0A212FC99
F4X4J6
+ More
A0A151JV17 A0A195BCU5 A0A158NIK6 A0A151XG45 A0A195CNP1 A0A232ELJ3 E2BFD6 K7J8Y7 A0A1W4XHW0 A0A0C9QGH1 A0A154P7A7 A0A3L8DNL8 A0A0J7KXI3 A0A0M8ZSV3 A0A026W231 A0A2P1DM47 A0A2J7R7F0 A0A2I6PIZ4 A0A0L7R5L2 A0A2A3EGM8 A0A088ACD3 A0A1Y1K9T9 A0A151J2X3 A0A310SU72 A0A067QJY1 A0A182T4I3 E9I8E2 A0A182Y052 A0A182U222 A0A182V1W0 A0A182WU62 A0A182J9R8 A0A1S4GX90 A0A182JTQ4 A0A182HUS4 Q5TQK8 A0A084VN71 A0A182L1K9 A0A182W763 A0A2R7VTY1 A0A182NUA6 A0A182PMM7 A0A182MAP4 A0A182QBA2 A0A182RTC0 A0A2P1DM88 A0A1Q3G295 A0A1B6C2Y5 A0A1S4J407 W5JRG0 A0A336LWE9 A0A336LTC6 A0A336MRR9 A0A1B0G5S3 A0A1A9US91 A0A0L0CPI3 A0A2M4A4G9 A0A182H177 A0A1A9ZAW6 A0A1A9XIV2 A0A1B0BUH8 Q16YA9 A0A1J1IPR8 A0A0K8V217 A0A182FJS9 A0A034W5R4 A0A023F4I7 A0A0A1XKY5 A0A224XDC0 A0A146L6Y6 W8CEJ8 B4KQ41 A0A1A9WEI4 A0A146KXR5 A0A0A9X7Y5 K7YZ60 T1I9Z1 B4J860 A0A0M3QVP3 B4NMS8 B4MF95 B3N9C5 A0A1I8NIB3 A0A3B0JPJ8 A0A3B0JJL1 A0A139WAR5 B4GD51 E0VZ53 Q28YB7 A0A1I8P8E5 B4P256 A0A1W4VZ00
A0A151JV17 A0A195BCU5 A0A158NIK6 A0A151XG45 A0A195CNP1 A0A232ELJ3 E2BFD6 K7J8Y7 A0A1W4XHW0 A0A0C9QGH1 A0A154P7A7 A0A3L8DNL8 A0A0J7KXI3 A0A0M8ZSV3 A0A026W231 A0A2P1DM47 A0A2J7R7F0 A0A2I6PIZ4 A0A0L7R5L2 A0A2A3EGM8 A0A088ACD3 A0A1Y1K9T9 A0A151J2X3 A0A310SU72 A0A067QJY1 A0A182T4I3 E9I8E2 A0A182Y052 A0A182U222 A0A182V1W0 A0A182WU62 A0A182J9R8 A0A1S4GX90 A0A182JTQ4 A0A182HUS4 Q5TQK8 A0A084VN71 A0A182L1K9 A0A182W763 A0A2R7VTY1 A0A182NUA6 A0A182PMM7 A0A182MAP4 A0A182QBA2 A0A182RTC0 A0A2P1DM88 A0A1Q3G295 A0A1B6C2Y5 A0A1S4J407 W5JRG0 A0A336LWE9 A0A336LTC6 A0A336MRR9 A0A1B0G5S3 A0A1A9US91 A0A0L0CPI3 A0A2M4A4G9 A0A182H177 A0A1A9ZAW6 A0A1A9XIV2 A0A1B0BUH8 Q16YA9 A0A1J1IPR8 A0A0K8V217 A0A182FJS9 A0A034W5R4 A0A023F4I7 A0A0A1XKY5 A0A224XDC0 A0A146L6Y6 W8CEJ8 B4KQ41 A0A1A9WEI4 A0A146KXR5 A0A0A9X7Y5 K7YZ60 T1I9Z1 B4J860 A0A0M3QVP3 B4NMS8 B4MF95 B3N9C5 A0A1I8NIB3 A0A3B0JPJ8 A0A3B0JJL1 A0A139WAR5 B4GD51 E0VZ53 Q28YB7 A0A1I8P8E5 B4P256 A0A1W4VZ00
PDB
5B16
E-value=2.92064e-168,
Score=1524
Ontologies
GO
Topology
Subcellular location
Membrane
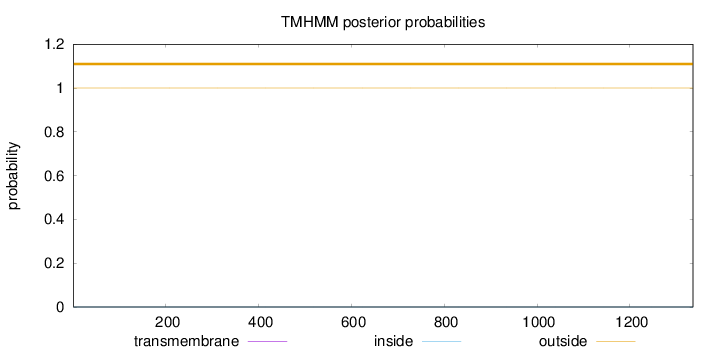
Length:
1336
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0016
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00001
outside
1 - 1336
Population Genetic Test Statistics
Pi
242.929034
Theta
182.44861
Tajima's D
1.118203
CLR
0.04393
CSRT
0.685265736713164
Interpretation
Uncertain
