Gene
KWMTBOMO10932
Pre Gene Modal
BGIBMGA008496
Annotation
PREDICTED:_uncharacterized_protein_LOC101745871_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.379
Sequence
CDS
ATGGACTTAGTAAATATACAGGTACTAGATCAAATAATAGAGTATTTATCGAAAATAGTTAAATCATATTATAAATTAAGTCCAAAGTTAGCGAGATTGAATAAAGATAGTGAATTGCTTCGAAACAAGCACGATCTTATTTTTGATAAACTCTGCAGTAGTAGATTTATAATGTTGCAAGCTATTCATGGACTTATAACGACATTACTAAACAACTGCTTGCACCGCGAGGACTCTAGGAAGATAATAGAATCGTGTTTGAATTTAGTCACAGTCTACAATCATCTTCTTGGCATAAGTTACAAAAAATTCAAAGCGAAATTCAACATATGCAGTAACACGTATCCAAATACTATTAGTGAAACAAAACCAATTACTTTGACCAAAATTGTACAGGTCTTGTCGAAGAGTCGTGTAGAAATTAATTGTCAACGACTTATAAATTGCTTAGTTAAGGCTTATAAGCCGCAAGAAGATTCCGATAACGACTCTAGTTCCAGTTCAGTGGACAGTTTAGAAATTTATCATACTTTAACCAAACACATAACGCCGCCATCGTCTTCCGACCGGAGAGGTTCAAATGAAAAACAGATAGTGAGCGGACAGAACTCTGACACCGATGTGCCTTATACCTCGCTAGTAAACAACGCACACGCTTTAGAAAATATTTTTAATAATGAAGATAATGTCAATAGTATTCTACAGAGCGAAGAAATGTTTATTGAAATATTATTAGATACTGTTTCGAATAAGTTTCCAACCTTACTTGGCCCCAATGGAGTAAAAAGATTGAAAACAGGTAGACTACAGGCCAGCAAAAAAGCTGCCCAAAAAGTAACCGAGTATTATCAAAATATTCTATGGGGTGATGTAGGAAGCTGTCTAGAACATATCGTCTTGTGGTGGTCTGCGTATCCCTTAGCTATGAGGTCCCCAAATACAAGTCAAAATCTTCGAGAATGGTTATTTAGCACTTTTAACATAAACAATACCCCAGATCTAATACTGTCTGCTTTGAGGATATTGGCCGATGCTTTGGGCTGCCATGTCACATCCACTTCTTGGGACAAAAATTTTGGTACTACTTTGCGGACCGATTTAACGCCAACAAACTTGAAACTTTATCCGGACCTTTCCTTGTACGTATCTGAGTGTACTGTGTCTGGTGCTAATTTCGCTCTAATGCTACAAGAATTAGTAACACTCAATAATCAGTGTGAGGTGACATGGGATTTTATATTGGGAGCGCCGATCGAGGATCTACCTATCGTCGAGCAAATACCCATTTTACATAGGCTTGATCATTCCATTCACACTTACCGGTTATGGTGCCTTGCGGTCACCCGCCGATTAGCCAATATGTGGGAGATGGATGATTTCTTCCGTGTCGCACACAATGACATGCAAGCTTGTATGGAACAATTACTTAACCTGAGATTCGCCGATCATTCGGCAACTATTGAAGAAGGGAAGATTGGGGTACATGAAAACGTTTGTGCAAAAATGAGAGAAAAATTAGTGTCCGAAGTGAAAGCCAATATACAGAAATTGAAGGAAGCTTCAGAAGAAATAACGCAGATGGTGTGCGCTGTTTGTAGTACGACAAGTTTAGCGCACTTGACGCGTATATTCCCGCATTCGTCGCACTGGAAGCGGCTCACTAATCCCCCTCAGCAGCACCACTCCCTATACGTACACTATTATCTGGATAAAATGCTACTGCCTGTGATTAAGGCAACTCAGGACGTCACCACACTAAACATGGTATTAAGAATTATGGGAGAGTCTTGGCTGCATCATATATACATGGCCAAAGTGAAATTTAGTAAAGATGCTGCTCAGCAACTCCTTGCTGATTTTAACGAGGTTCGAAACTGGTTGAATGATTGTAAGCTCCTAACGGTCGCGGCTAGGAAACAAATGTTAAATAACGAAGTTTTAAGACGATGTGAAGGTGTCGGCCGCCTGCTATTACATGCACCAGGCGATCTTATATCTATGCAGGAGTCTACTATGCAGTCTGCCCAACAAATCCGGAAAGATGTAACAGAAAACGATTCTCCAGAGCAAATGATGCCGGCGGAGATGTATGTACCGAACCAGAAACAGTGGCTCACTCTCAGAGCGGGAAAGCTGAAAGGACCTGTAGTTTTTTCTCTGTGCTGTGTCGTATTATTGTGA
Protein
MDLVNIQVLDQIIEYLSKIVKSYYKLSPKLARLNKDSELLRNKHDLIFDKLCSSRFIMLQAIHGLITTLLNNCLHREDSRKIIESCLNLVTVYNHLLGISYKKFKAKFNICSNTYPNTISETKPITLTKIVQVLSKSRVEINCQRLINCLVKAYKPQEDSDNDSSSSSVDSLEIYHTLTKHITPPSSSDRRGSNEKQIVSGQNSDTDVPYTSLVNNAHALENIFNNEDNVNSILQSEEMFIEILLDTVSNKFPTLLGPNGVKRLKTGRLQASKKAAQKVTEYYQNILWGDVGSCLEHIVLWWSAYPLAMRSPNTSQNLREWLFSTFNINNTPDLILSALRILADALGCHVTSTSWDKNFGTTLRTDLTPTNLKLYPDLSLYVSECTVSGANFALMLQELVTLNNQCEVTWDFILGAPIEDLPIVEQIPILHRLDHSIHTYRLWCLAVTRRLANMWEMDDFFRVAHNDMQACMEQLLNLRFADHSATIEEGKIGVHENVCAKMREKLVSEVKANIQKLKEASEEITQMVCAVCSTTSLAHLTRIFPHSSHWKRLTNPPQQHHSLYVHYYLDKMLLPVIKATQDVTTLNMVLRIMGESWLHHIYMAKVKFSKDAAQQLLADFNEVRNWLNDCKLLTVAARKQMLNNEVLRRCEGVGRLLLHAPGDLISMQESTMQSAQQIRKDVTENDSPEQMMPAEMYVPNQKQWLTLRAGKLKGPVVFSLCCVVLL
Summary
Uniprot
H9JG49
A0A2H1VJ63
A0A2A4K4R6
A0A194PIL4
A0A212FCA0
A0A194RV84
+ More
D6X317 A0A1W4XEX4 E2BYM4 A0A0M9A9U3 F4X4A1 A0A151WEA9 A0A158NBW8 A0A195E4I7 A0A195FHP3 A0A088AGC9 A0A195BDU8 E9IAK3 A0A0L7RJ29 A0A2A3EEG7 A0A310SSD5 A0A2J7PBB6 A0A026X3E3 A0A2P8Y6W1 A0A1W4XPJ0 A0A1B6E8U2 E2A2Y3 A0A2J7PBD4 A0A232FJS4 W5JTR2 A0A084WT42 A0A182T7K9 A0A182F7X1 A0A182M5R5 A0A1S4FYU2 A0A182UHA5 A0A182HD69 A0A182K4Z2 Q16JI5 A0A182Y4M2 A0A182KWT7 A0A146KUE0 A0A182NST2 A0A182VRW8 A0A182XKB6 A0A182HJM7 A0A182RH07 A0A182QKV8 A0A182PCW1 A0A1B0C9X3 B0W4D7 E0VZQ0 A0A336MAF7 Q29AT6 B4G5V1 T1HAZ7 A0A3B0JKY8 A0A1I8PN77 B4N9Q8 A0A1I8MZP7 B3LZU2 A0A0R3NKT2 Q9VCV8 A0A1W4VLS8 B4KAC4 B4PME2 A0A0L0CF94 A0A1J1HRJ7 A0A0M4EN32 B3P8K8 B4JIE5 B4LXC6 A0A1B0BNM6 A0A1B0G7Y2 A0A3L8DJX3 A0A1A9XGA7 A0A1A9ZHN7 B4R0P3 A0A1J1HTL2
D6X317 A0A1W4XEX4 E2BYM4 A0A0M9A9U3 F4X4A1 A0A151WEA9 A0A158NBW8 A0A195E4I7 A0A195FHP3 A0A088AGC9 A0A195BDU8 E9IAK3 A0A0L7RJ29 A0A2A3EEG7 A0A310SSD5 A0A2J7PBB6 A0A026X3E3 A0A2P8Y6W1 A0A1W4XPJ0 A0A1B6E8U2 E2A2Y3 A0A2J7PBD4 A0A232FJS4 W5JTR2 A0A084WT42 A0A182T7K9 A0A182F7X1 A0A182M5R5 A0A1S4FYU2 A0A182UHA5 A0A182HD69 A0A182K4Z2 Q16JI5 A0A182Y4M2 A0A182KWT7 A0A146KUE0 A0A182NST2 A0A182VRW8 A0A182XKB6 A0A182HJM7 A0A182RH07 A0A182QKV8 A0A182PCW1 A0A1B0C9X3 B0W4D7 E0VZQ0 A0A336MAF7 Q29AT6 B4G5V1 T1HAZ7 A0A3B0JKY8 A0A1I8PN77 B4N9Q8 A0A1I8MZP7 B3LZU2 A0A0R3NKT2 Q9VCV8 A0A1W4VLS8 B4KAC4 B4PME2 A0A0L0CF94 A0A1J1HRJ7 A0A0M4EN32 B3P8K8 B4JIE5 B4LXC6 A0A1B0BNM6 A0A1B0G7Y2 A0A3L8DJX3 A0A1A9XGA7 A0A1A9ZHN7 B4R0P3 A0A1J1HTL2
Pubmed
19121390
26354079
22118469
18362917
19820115
20798317
+ More
21719571 21347285 21282665 24508170 29403074 28648823 20920257 23761445 24438588 26483478 17510324 25244985 20966253 26823975 20566863 15632085 17994087 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 26108605 30249741
21719571 21347285 21282665 24508170 29403074 28648823 20920257 23761445 24438588 26483478 17510324 25244985 20966253 26823975 20566863 15632085 17994087 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 26108605 30249741
EMBL
BABH01001985
ODYU01002848
SOQ40836.1
NWSH01000165
PCG78904.1
KQ459604
+ More
KPI92559.1 AGBW02009208 OWR51364.1 KQ459765 KPJ20076.1 KQ971372 EFA09809.1 GL451499 EFN79178.1 KQ435706 KOX80150.1 GL888633 EGI58697.1 KQ983238 KYQ46199.1 ADTU01000447 ADTU01000448 KQ979685 KYN19817.1 KQ981606 KYN39524.1 KQ976509 KYM82733.1 GL762022 EFZ22455.1 KQ414581 KOC70967.1 KZ288271 PBC29864.1 KQ759821 OAD62687.1 NEVH01027114 PNF13636.1 KK107019 EZA62782.1 PYGN01000865 PSN39904.1 GEDC01002949 JAS34349.1 GL436239 EFN72191.1 PNF13644.1 NNAY01000115 OXU30833.1 ADMH02000406 ETN66673.1 ATLV01026830 KE525419 KFB53386.1 AXCM01012226 JXUM01129063 JXUM01129064 JXUM01129065 JXUM01129066 KQ567396 KXJ69453.1 CH478007 EAT34440.1 GDHC01019733 JAP98895.1 APCN01002191 AXCN02000300 AJWK01002981 DS231836 EDS33222.1 DS235854 EEB18856.1 UFQT01000789 SSX27246.1 CM000070 EAL27263.1 CH479179 EDW23744.1 ACPB03019361 OUUW01000007 SPP82987.1 CH964232 EDW80623.1 CH902617 EDV43086.1 KRS99892.1 AE014297 AAF56047.2 CH933806 EDW14611.1 CM000160 EDW98047.2 JRES01000563 KNC30174.1 CVRI01000020 CRK90603.1 CP012526 ALC46001.1 CH954182 EDV54103.1 CH916369 EDV93026.1 CH940650 EDW67804.1 JXJN01017478 CCAG010010575 CCAG010010576 CCAG010010577 QOIP01000007 RLU20473.1 CM000364 EDX13966.1 CRK91331.1
KPI92559.1 AGBW02009208 OWR51364.1 KQ459765 KPJ20076.1 KQ971372 EFA09809.1 GL451499 EFN79178.1 KQ435706 KOX80150.1 GL888633 EGI58697.1 KQ983238 KYQ46199.1 ADTU01000447 ADTU01000448 KQ979685 KYN19817.1 KQ981606 KYN39524.1 KQ976509 KYM82733.1 GL762022 EFZ22455.1 KQ414581 KOC70967.1 KZ288271 PBC29864.1 KQ759821 OAD62687.1 NEVH01027114 PNF13636.1 KK107019 EZA62782.1 PYGN01000865 PSN39904.1 GEDC01002949 JAS34349.1 GL436239 EFN72191.1 PNF13644.1 NNAY01000115 OXU30833.1 ADMH02000406 ETN66673.1 ATLV01026830 KE525419 KFB53386.1 AXCM01012226 JXUM01129063 JXUM01129064 JXUM01129065 JXUM01129066 KQ567396 KXJ69453.1 CH478007 EAT34440.1 GDHC01019733 JAP98895.1 APCN01002191 AXCN02000300 AJWK01002981 DS231836 EDS33222.1 DS235854 EEB18856.1 UFQT01000789 SSX27246.1 CM000070 EAL27263.1 CH479179 EDW23744.1 ACPB03019361 OUUW01000007 SPP82987.1 CH964232 EDW80623.1 CH902617 EDV43086.1 KRS99892.1 AE014297 AAF56047.2 CH933806 EDW14611.1 CM000160 EDW98047.2 JRES01000563 KNC30174.1 CVRI01000020 CRK90603.1 CP012526 ALC46001.1 CH954182 EDV54103.1 CH916369 EDV93026.1 CH940650 EDW67804.1 JXJN01017478 CCAG010010575 CCAG010010576 CCAG010010577 QOIP01000007 RLU20473.1 CM000364 EDX13966.1 CRK91331.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000007266
+ More
UP000192223 UP000008237 UP000053105 UP000007755 UP000075809 UP000005205 UP000078492 UP000078541 UP000005203 UP000078540 UP000053825 UP000242457 UP000235965 UP000053097 UP000245037 UP000000311 UP000215335 UP000000673 UP000030765 UP000075901 UP000069272 UP000075883 UP000075902 UP000069940 UP000249989 UP000075881 UP000008820 UP000076408 UP000075882 UP000075884 UP000075920 UP000076407 UP000075840 UP000075900 UP000075886 UP000075885 UP000092461 UP000002320 UP000009046 UP000001819 UP000008744 UP000015103 UP000268350 UP000095300 UP000007798 UP000095301 UP000007801 UP000000803 UP000192221 UP000009192 UP000002282 UP000037069 UP000183832 UP000092553 UP000008711 UP000001070 UP000008792 UP000092460 UP000092444 UP000279307 UP000092443 UP000092445 UP000000304
UP000192223 UP000008237 UP000053105 UP000007755 UP000075809 UP000005205 UP000078492 UP000078541 UP000005203 UP000078540 UP000053825 UP000242457 UP000235965 UP000053097 UP000245037 UP000000311 UP000215335 UP000000673 UP000030765 UP000075901 UP000069272 UP000075883 UP000075902 UP000069940 UP000249989 UP000075881 UP000008820 UP000076408 UP000075882 UP000075884 UP000075920 UP000076407 UP000075840 UP000075900 UP000075886 UP000075885 UP000092461 UP000002320 UP000009046 UP000001819 UP000008744 UP000015103 UP000268350 UP000095300 UP000007798 UP000095301 UP000007801 UP000000803 UP000192221 UP000009192 UP000002282 UP000037069 UP000183832 UP000092553 UP000008711 UP000001070 UP000008792 UP000092460 UP000092444 UP000279307 UP000092443 UP000092445 UP000000304
Interpro
SUPFAM
SSF47473
SSF47473
ProteinModelPortal
H9JG49
A0A2H1VJ63
A0A2A4K4R6
A0A194PIL4
A0A212FCA0
A0A194RV84
+ More
D6X317 A0A1W4XEX4 E2BYM4 A0A0M9A9U3 F4X4A1 A0A151WEA9 A0A158NBW8 A0A195E4I7 A0A195FHP3 A0A088AGC9 A0A195BDU8 E9IAK3 A0A0L7RJ29 A0A2A3EEG7 A0A310SSD5 A0A2J7PBB6 A0A026X3E3 A0A2P8Y6W1 A0A1W4XPJ0 A0A1B6E8U2 E2A2Y3 A0A2J7PBD4 A0A232FJS4 W5JTR2 A0A084WT42 A0A182T7K9 A0A182F7X1 A0A182M5R5 A0A1S4FYU2 A0A182UHA5 A0A182HD69 A0A182K4Z2 Q16JI5 A0A182Y4M2 A0A182KWT7 A0A146KUE0 A0A182NST2 A0A182VRW8 A0A182XKB6 A0A182HJM7 A0A182RH07 A0A182QKV8 A0A182PCW1 A0A1B0C9X3 B0W4D7 E0VZQ0 A0A336MAF7 Q29AT6 B4G5V1 T1HAZ7 A0A3B0JKY8 A0A1I8PN77 B4N9Q8 A0A1I8MZP7 B3LZU2 A0A0R3NKT2 Q9VCV8 A0A1W4VLS8 B4KAC4 B4PME2 A0A0L0CF94 A0A1J1HRJ7 A0A0M4EN32 B3P8K8 B4JIE5 B4LXC6 A0A1B0BNM6 A0A1B0G7Y2 A0A3L8DJX3 A0A1A9XGA7 A0A1A9ZHN7 B4R0P3 A0A1J1HTL2
D6X317 A0A1W4XEX4 E2BYM4 A0A0M9A9U3 F4X4A1 A0A151WEA9 A0A158NBW8 A0A195E4I7 A0A195FHP3 A0A088AGC9 A0A195BDU8 E9IAK3 A0A0L7RJ29 A0A2A3EEG7 A0A310SSD5 A0A2J7PBB6 A0A026X3E3 A0A2P8Y6W1 A0A1W4XPJ0 A0A1B6E8U2 E2A2Y3 A0A2J7PBD4 A0A232FJS4 W5JTR2 A0A084WT42 A0A182T7K9 A0A182F7X1 A0A182M5R5 A0A1S4FYU2 A0A182UHA5 A0A182HD69 A0A182K4Z2 Q16JI5 A0A182Y4M2 A0A182KWT7 A0A146KUE0 A0A182NST2 A0A182VRW8 A0A182XKB6 A0A182HJM7 A0A182RH07 A0A182QKV8 A0A182PCW1 A0A1B0C9X3 B0W4D7 E0VZQ0 A0A336MAF7 Q29AT6 B4G5V1 T1HAZ7 A0A3B0JKY8 A0A1I8PN77 B4N9Q8 A0A1I8MZP7 B3LZU2 A0A0R3NKT2 Q9VCV8 A0A1W4VLS8 B4KAC4 B4PME2 A0A0L0CF94 A0A1J1HRJ7 A0A0M4EN32 B3P8K8 B4JIE5 B4LXC6 A0A1B0BNM6 A0A1B0G7Y2 A0A3L8DJX3 A0A1A9XGA7 A0A1A9ZHN7 B4R0P3 A0A1J1HTL2
Ontologies
PANTHER
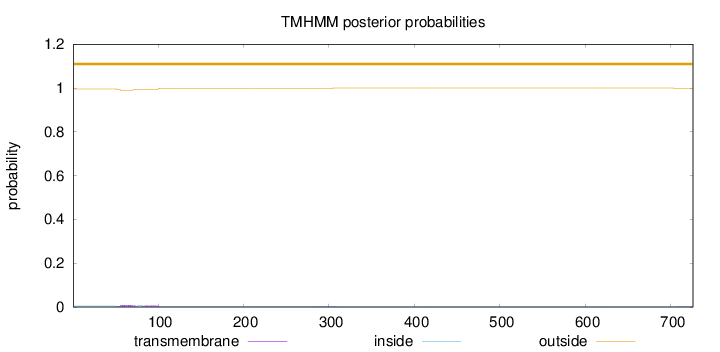
Topology
Length:
726
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.313520000000001
Exp number, first 60 AAs:
0.06169
Total prob of N-in:
0.00480
outside
1 - 726
Population Genetic Test Statistics
Pi
197.406228
Theta
158.384199
Tajima's D
0.634115
CLR
0.051434
CSRT
0.550072496375181
Interpretation
Uncertain
