Gene
KWMTBOMO10931
Pre Gene Modal
BGIBMGA008495
Annotation
PREDICTED:_transmembrane_protein_185B_isoform_X2_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 4.989
Sequence
CDS
ATGAATTTACAGACATTATTTCAAGACTTTAACCCCAGCAAATTCGTCATTCACACTTGTCTTCTAATATTTACTGCATTATTAGCGCTGAAGTTAGATAAAACCATAACATGGTCATATTGGGCAGTATTTACTCCAATATGGATTTGGAAATTTCTTGTCGTGCTCGGGGCCACTATTGGGACTTATGTGTGGTGGCAGTATCCTCACTTCAGATTGGAGGGAGAAGCATACATCCATTATAAAGCTATGCTGATAAGTCTTGCCATTCACCTAATATTACTTATGTTTGAGTTACTTGTCTGTGACCAGTTAACTACCAGCCGCCATGTATGGATCCTAGTCTTTATACCTTTAATCTTTATAAGTATAGTTTCCATTGCCATCTGTATATGGTCAGTAAAACATGATCGAAGTTATGAGTTGGAATTATTTTGTGCAGTGAATATACTTCAGTTTATATTCTTGGCCTTGAAATTGGATAAATTTATACACTGGTCCTGGGAGGTTGTGTTTGTCCCACTATGGATATTAATGTGCCTCAGCCTTGTAGGTGTTTTATACAGTATCATATTTGCTGGAATTTTACTACGAACTCCCGAAGTTAACGTACAGCAGAGACGCACAAGCTTCAACTCAGCACTTGCCTACACTTTCACTGTGATACCTATCTTGGTATTCCAGGTTCTATTAACTAACAAGTTAGATGATGGCCTATCATTTACTTATATAGTGATAGTGAGTCCGCTATTCATCAGTTATGCCACACTAATTATTATGTCGTTCAGTTCAAAAGGAGGAAACAAATGGTGGTTTGGTATCCGAAAAGATTTCTGTCAATTCTTGTTATCCGTATTTCCACTGTTGCAAGAATACGCGAATATCGCTTACAACAACAATAACGTGAACCGCTCTAACGGGATCCCGAACGAGCTTCCCGCGAACCAGGAGCCGTACAGAGATGAGGAGACCCACAAAAAAGTCAAAGAGAACAAGAGATCGGCTAAAAATCAGAAAAAAAACGAAGCTATGAAACCGGTCGTGCCTATTACCAGCATAGATATGCCGGACTGA
Protein
MNLQTLFQDFNPSKFVIHTCLLIFTALLALKLDKTITWSYWAVFTPIWIWKFLVVLGATIGTYVWWQYPHFRLEGEAYIHYKAMLISLAIHLILLMFELLVCDQLTTSRHVWILVFIPLIFISIVSIAICIWSVKHDRSYELELFCAVNILQFIFLALKLDKFIHWSWEVVFVPLWILMCLSLVGVLYSIIFAGILLRTPEVNVQQRRTSFNSALAYTFTVIPILVFQVLLTNKLDDGLSFTYIVIVSPLFISYATLIIMSFSSKGGNKWWFGIRKDFCQFLLSVFPLLQEYANIAYNNNNVNRSNGIPNELPANQEPYRDEETHKKVKENKRSAKNQKKNEAMKPVVPITSIDMPD
Summary
Uniprot
A0A3S2L6A3
A0A1E1WAG5
A0A2W1BVW1
A0A2A4K557
A0A194PN66
A0A212FCB8
+ More
A0A2J7PM42 A0A1B6F6R2 A0A1B6H5F8 A0A1B6L054 A0A1B6LJH7 A0A146M5F1 A0A0A9W8D9 A0A067RBQ6 A0A2H1WQ78 N6UG20 A0A0P4VWH6 A0A0V0G9C6 A0A023FAV6 A0A069DSC7 R4G8R4 J3JXM7 E0VPU1 A0A1Y1KNH6 A0A1W4X8F3 A0A1B6E3D8 A0A194RRI3 K7J203 E2B9J6 F4WM16 A0A195FUT5 A0A151XA49 E2AK96 A0A158N951 A0A195C831 A0A026W2D9 A0A0C9RDG2 A0A088AD08 A0A2A3ELX5 A0A195DCI7 A0A195BC39 A0A232ELR2 A0A0M8ZUR8 A0A0L7KGT5 B4NBZ0 A0A1B6C868 B3N152 Q29GQ1 A0A3B0KRX0 A0A2M3Z4M0 A0A2M4AGS3 A0A2M4BSP2 A0A0N8ES82 Q9VWK7 B4R7B9 B4I722 A0A182QR00 A0A0P4WGC5 B4PWT1 A0A023ERI6 A0A0L7QWX3 A0A1D2N5H8 U5EWF6 A0A1A9WVE5 A0A1B0FIW4 A0A1A9VV96 A0A1A9Z9R6 A0A1A9YQN2 B3NUS8 W5JA08 A0A182NQZ3 A0A1W4V2V4 A0A182YNC7 A0A0K8TY60 A0A034WG65 A0A182RFZ9 A0A1W7R821 A0A182INP6 L7M7Y3 W8BY11 A0A182JRG2 A0A224YJL3 A0A131YXT4 B4JNN4 A0A2T7PT74 B0WP84 A0A1I8N4A6 A0A1E1X4I3 A0A023GM14 G3MRD4 A0A1Q3FGU2 A0A2R5LHF6 A0A1I8QEZ5 A0A1Q3FGW4 A0A131Y4X7 V4A6H8 T1IRI0 T1E842 A0A293M5Z7
A0A2J7PM42 A0A1B6F6R2 A0A1B6H5F8 A0A1B6L054 A0A1B6LJH7 A0A146M5F1 A0A0A9W8D9 A0A067RBQ6 A0A2H1WQ78 N6UG20 A0A0P4VWH6 A0A0V0G9C6 A0A023FAV6 A0A069DSC7 R4G8R4 J3JXM7 E0VPU1 A0A1Y1KNH6 A0A1W4X8F3 A0A1B6E3D8 A0A194RRI3 K7J203 E2B9J6 F4WM16 A0A195FUT5 A0A151XA49 E2AK96 A0A158N951 A0A195C831 A0A026W2D9 A0A0C9RDG2 A0A088AD08 A0A2A3ELX5 A0A195DCI7 A0A195BC39 A0A232ELR2 A0A0M8ZUR8 A0A0L7KGT5 B4NBZ0 A0A1B6C868 B3N152 Q29GQ1 A0A3B0KRX0 A0A2M3Z4M0 A0A2M4AGS3 A0A2M4BSP2 A0A0N8ES82 Q9VWK7 B4R7B9 B4I722 A0A182QR00 A0A0P4WGC5 B4PWT1 A0A023ERI6 A0A0L7QWX3 A0A1D2N5H8 U5EWF6 A0A1A9WVE5 A0A1B0FIW4 A0A1A9VV96 A0A1A9Z9R6 A0A1A9YQN2 B3NUS8 W5JA08 A0A182NQZ3 A0A1W4V2V4 A0A182YNC7 A0A0K8TY60 A0A034WG65 A0A182RFZ9 A0A1W7R821 A0A182INP6 L7M7Y3 W8BY11 A0A182JRG2 A0A224YJL3 A0A131YXT4 B4JNN4 A0A2T7PT74 B0WP84 A0A1I8N4A6 A0A1E1X4I3 A0A023GM14 G3MRD4 A0A1Q3FGU2 A0A2R5LHF6 A0A1I8QEZ5 A0A1Q3FGW4 A0A131Y4X7 V4A6H8 T1IRI0 T1E842 A0A293M5Z7
Pubmed
28756777
26354079
22118469
26823975
25401762
24845553
+ More
23537049 27129103 25474469 26334808 22516182 20566863 28004739 20075255 20798317 21719571 21347285 24508170 30249741 28648823 26227816 17994087 15632085 26999592 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 24945155 27289101 20920257 23761445 25244985 25348373 25576852 24495485 28797301 26830274 25315136 28503490 22216098 29652888 23254933
23537049 27129103 25474469 26334808 22516182 20566863 28004739 20075255 20798317 21719571 21347285 24508170 30249741 28648823 26227816 17994087 15632085 26999592 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 24945155 27289101 20920257 23761445 25244985 25348373 25576852 24495485 28797301 26830274 25315136 28503490 22216098 29652888 23254933
EMBL
RSAL01000123
RVE46638.1
GDQN01007098
JAT83956.1
KZ149920
PZC77764.1
+ More
NWSH01000165 PCG78903.1 KQ459604 KPI92560.1 AGBW02009208 OWR51363.1 NEVH01024421 PNF17408.1 GECZ01023925 JAS45844.1 GECU01037767 JAS69939.1 GEBQ01022880 JAT17097.1 GEBQ01016157 JAT23820.1 GDHC01003970 JAQ14659.1 GBHO01039545 JAG04059.1 KK852807 KDR16080.1 ODYU01010245 SOQ55225.1 APGK01036620 KB740941 KB632409 ENN77582.1 ERL95166.1 GDKW01000117 JAI56478.1 GECL01001354 JAP04770.1 GBBI01000654 JAC18058.1 GBGD01001921 JAC86968.1 GAHY01000517 JAA76993.1 BT127997 AEE62959.1 DS235379 EEB15397.1 GEZM01078212 GEZM01078209 JAV62854.1 GEDC01004873 JAS32425.1 KQ459765 KPJ20077.1 GL446556 EFN87629.1 GL888217 EGI64660.1 KQ981264 KYN44052.1 KQ982353 KYQ57180.1 GL440209 EFN66137.1 ADTU01001366 KQ978220 KYM96326.1 KK107474 QOIP01000002 EZA50163.1 RLU25612.1 GBYB01006302 GBYB01006305 GBYB01006306 GBYB01006856 JAG76069.1 JAG76072.1 JAG76073.1 JAG76623.1 KZ288220 PBC32169.1 KQ980989 KYN10625.1 KQ976519 KYM82131.1 NNAY01003514 OXU19295.1 KQ435878 KOX70023.1 JTDY01009835 KOB62542.1 CH964239 EDW82349.1 GEDC01027605 JAS09693.1 CH902650 EDV30087.1 CH379064 EAL32058.1 OUUW01000015 SPP88646.1 GGFM01002694 MBW23445.1 GGFK01006659 MBW39980.1 GGFJ01006965 MBW56106.1 GDUN01000194 JAN95725.1 AE014298 AY119255 AAF48931.1 AAM51115.1 CM000366 EDX18360.1 CH480823 EDW56120.1 AXCN02001485 GDRN01015312 JAI67847.1 CM000162 EDX02815.1 GAPW01002314 JAC11284.1 KQ414706 KOC63095.1 LJIJ01000202 ODN00517.1 GANO01000600 JAB59271.1 CCAG010016654 CH954180 EDV46675.1 ADMH02002066 ETN59690.1 GDHF01033108 GDHF01016679 GDHF01009767 GDHF01005090 JAI19206.1 JAI35635.1 JAI42547.1 JAI47224.1 GAKP01005643 GAKP01005640 JAC53312.1 GEHC01000359 JAV47286.1 GACK01004628 JAA60406.1 GAMC01002318 JAC04238.1 GFPF01006691 MAA17837.1 GEDV01004770 JAP83787.1 CH916371 EDV92327.1 PZQS01000002 PVD36626.1 DS232020 EDS32129.1 GFAC01005023 JAT94165.1 GBBM01001553 JAC33865.1 JO844435 AEO36052.1 GFDL01008291 JAV26754.1 GGLE01004729 MBY08855.1 GFDL01008276 JAV26769.1 GEFM01002690 GEGO01002434 JAP73106.1 JAR92970.1 KB202620 ESO88851.1 JH431361 GAMD01002517 JAA99073.1 GFWV01011664 MAA36393.1
NWSH01000165 PCG78903.1 KQ459604 KPI92560.1 AGBW02009208 OWR51363.1 NEVH01024421 PNF17408.1 GECZ01023925 JAS45844.1 GECU01037767 JAS69939.1 GEBQ01022880 JAT17097.1 GEBQ01016157 JAT23820.1 GDHC01003970 JAQ14659.1 GBHO01039545 JAG04059.1 KK852807 KDR16080.1 ODYU01010245 SOQ55225.1 APGK01036620 KB740941 KB632409 ENN77582.1 ERL95166.1 GDKW01000117 JAI56478.1 GECL01001354 JAP04770.1 GBBI01000654 JAC18058.1 GBGD01001921 JAC86968.1 GAHY01000517 JAA76993.1 BT127997 AEE62959.1 DS235379 EEB15397.1 GEZM01078212 GEZM01078209 JAV62854.1 GEDC01004873 JAS32425.1 KQ459765 KPJ20077.1 GL446556 EFN87629.1 GL888217 EGI64660.1 KQ981264 KYN44052.1 KQ982353 KYQ57180.1 GL440209 EFN66137.1 ADTU01001366 KQ978220 KYM96326.1 KK107474 QOIP01000002 EZA50163.1 RLU25612.1 GBYB01006302 GBYB01006305 GBYB01006306 GBYB01006856 JAG76069.1 JAG76072.1 JAG76073.1 JAG76623.1 KZ288220 PBC32169.1 KQ980989 KYN10625.1 KQ976519 KYM82131.1 NNAY01003514 OXU19295.1 KQ435878 KOX70023.1 JTDY01009835 KOB62542.1 CH964239 EDW82349.1 GEDC01027605 JAS09693.1 CH902650 EDV30087.1 CH379064 EAL32058.1 OUUW01000015 SPP88646.1 GGFM01002694 MBW23445.1 GGFK01006659 MBW39980.1 GGFJ01006965 MBW56106.1 GDUN01000194 JAN95725.1 AE014298 AY119255 AAF48931.1 AAM51115.1 CM000366 EDX18360.1 CH480823 EDW56120.1 AXCN02001485 GDRN01015312 JAI67847.1 CM000162 EDX02815.1 GAPW01002314 JAC11284.1 KQ414706 KOC63095.1 LJIJ01000202 ODN00517.1 GANO01000600 JAB59271.1 CCAG010016654 CH954180 EDV46675.1 ADMH02002066 ETN59690.1 GDHF01033108 GDHF01016679 GDHF01009767 GDHF01005090 JAI19206.1 JAI35635.1 JAI42547.1 JAI47224.1 GAKP01005643 GAKP01005640 JAC53312.1 GEHC01000359 JAV47286.1 GACK01004628 JAA60406.1 GAMC01002318 JAC04238.1 GFPF01006691 MAA17837.1 GEDV01004770 JAP83787.1 CH916371 EDV92327.1 PZQS01000002 PVD36626.1 DS232020 EDS32129.1 GFAC01005023 JAT94165.1 GBBM01001553 JAC33865.1 JO844435 AEO36052.1 GFDL01008291 JAV26754.1 GGLE01004729 MBY08855.1 GFDL01008276 JAV26769.1 GEFM01002690 GEGO01002434 JAP73106.1 JAR92970.1 KB202620 ESO88851.1 JH431361 GAMD01002517 JAA99073.1 GFWV01011664 MAA36393.1
Proteomes
UP000283053
UP000218220
UP000053268
UP000007151
UP000235965
UP000027135
+ More
UP000019118 UP000030742 UP000009046 UP000192223 UP000053240 UP000002358 UP000008237 UP000007755 UP000078541 UP000075809 UP000000311 UP000005205 UP000078542 UP000053097 UP000279307 UP000005203 UP000242457 UP000078492 UP000078540 UP000215335 UP000053105 UP000037510 UP000007798 UP000007801 UP000001819 UP000268350 UP000000803 UP000000304 UP000001292 UP000075886 UP000002282 UP000053825 UP000094527 UP000091820 UP000092444 UP000078200 UP000092445 UP000092443 UP000008711 UP000000673 UP000075884 UP000192221 UP000076408 UP000075900 UP000075880 UP000075881 UP000001070 UP000245119 UP000002320 UP000095301 UP000095300 UP000030746
UP000019118 UP000030742 UP000009046 UP000192223 UP000053240 UP000002358 UP000008237 UP000007755 UP000078541 UP000075809 UP000000311 UP000005205 UP000078542 UP000053097 UP000279307 UP000005203 UP000242457 UP000078492 UP000078540 UP000215335 UP000053105 UP000037510 UP000007798 UP000007801 UP000001819 UP000268350 UP000000803 UP000000304 UP000001292 UP000075886 UP000002282 UP000053825 UP000094527 UP000091820 UP000092444 UP000078200 UP000092445 UP000092443 UP000008711 UP000000673 UP000075884 UP000192221 UP000076408 UP000075900 UP000075880 UP000075881 UP000001070 UP000245119 UP000002320 UP000095301 UP000095300 UP000030746
PRIDE
Pfam
PF10269 Tmemb_185A
Interpro
IPR019396
TM_Fragile-X-F-assoc
ProteinModelPortal
A0A3S2L6A3
A0A1E1WAG5
A0A2W1BVW1
A0A2A4K557
A0A194PN66
A0A212FCB8
+ More
A0A2J7PM42 A0A1B6F6R2 A0A1B6H5F8 A0A1B6L054 A0A1B6LJH7 A0A146M5F1 A0A0A9W8D9 A0A067RBQ6 A0A2H1WQ78 N6UG20 A0A0P4VWH6 A0A0V0G9C6 A0A023FAV6 A0A069DSC7 R4G8R4 J3JXM7 E0VPU1 A0A1Y1KNH6 A0A1W4X8F3 A0A1B6E3D8 A0A194RRI3 K7J203 E2B9J6 F4WM16 A0A195FUT5 A0A151XA49 E2AK96 A0A158N951 A0A195C831 A0A026W2D9 A0A0C9RDG2 A0A088AD08 A0A2A3ELX5 A0A195DCI7 A0A195BC39 A0A232ELR2 A0A0M8ZUR8 A0A0L7KGT5 B4NBZ0 A0A1B6C868 B3N152 Q29GQ1 A0A3B0KRX0 A0A2M3Z4M0 A0A2M4AGS3 A0A2M4BSP2 A0A0N8ES82 Q9VWK7 B4R7B9 B4I722 A0A182QR00 A0A0P4WGC5 B4PWT1 A0A023ERI6 A0A0L7QWX3 A0A1D2N5H8 U5EWF6 A0A1A9WVE5 A0A1B0FIW4 A0A1A9VV96 A0A1A9Z9R6 A0A1A9YQN2 B3NUS8 W5JA08 A0A182NQZ3 A0A1W4V2V4 A0A182YNC7 A0A0K8TY60 A0A034WG65 A0A182RFZ9 A0A1W7R821 A0A182INP6 L7M7Y3 W8BY11 A0A182JRG2 A0A224YJL3 A0A131YXT4 B4JNN4 A0A2T7PT74 B0WP84 A0A1I8N4A6 A0A1E1X4I3 A0A023GM14 G3MRD4 A0A1Q3FGU2 A0A2R5LHF6 A0A1I8QEZ5 A0A1Q3FGW4 A0A131Y4X7 V4A6H8 T1IRI0 T1E842 A0A293M5Z7
A0A2J7PM42 A0A1B6F6R2 A0A1B6H5F8 A0A1B6L054 A0A1B6LJH7 A0A146M5F1 A0A0A9W8D9 A0A067RBQ6 A0A2H1WQ78 N6UG20 A0A0P4VWH6 A0A0V0G9C6 A0A023FAV6 A0A069DSC7 R4G8R4 J3JXM7 E0VPU1 A0A1Y1KNH6 A0A1W4X8F3 A0A1B6E3D8 A0A194RRI3 K7J203 E2B9J6 F4WM16 A0A195FUT5 A0A151XA49 E2AK96 A0A158N951 A0A195C831 A0A026W2D9 A0A0C9RDG2 A0A088AD08 A0A2A3ELX5 A0A195DCI7 A0A195BC39 A0A232ELR2 A0A0M8ZUR8 A0A0L7KGT5 B4NBZ0 A0A1B6C868 B3N152 Q29GQ1 A0A3B0KRX0 A0A2M3Z4M0 A0A2M4AGS3 A0A2M4BSP2 A0A0N8ES82 Q9VWK7 B4R7B9 B4I722 A0A182QR00 A0A0P4WGC5 B4PWT1 A0A023ERI6 A0A0L7QWX3 A0A1D2N5H8 U5EWF6 A0A1A9WVE5 A0A1B0FIW4 A0A1A9VV96 A0A1A9Z9R6 A0A1A9YQN2 B3NUS8 W5JA08 A0A182NQZ3 A0A1W4V2V4 A0A182YNC7 A0A0K8TY60 A0A034WG65 A0A182RFZ9 A0A1W7R821 A0A182INP6 L7M7Y3 W8BY11 A0A182JRG2 A0A224YJL3 A0A131YXT4 B4JNN4 A0A2T7PT74 B0WP84 A0A1I8N4A6 A0A1E1X4I3 A0A023GM14 G3MRD4 A0A1Q3FGU2 A0A2R5LHF6 A0A1I8QEZ5 A0A1Q3FGW4 A0A131Y4X7 V4A6H8 T1IRI0 T1E842 A0A293M5Z7
Ontologies
GO
PANTHER
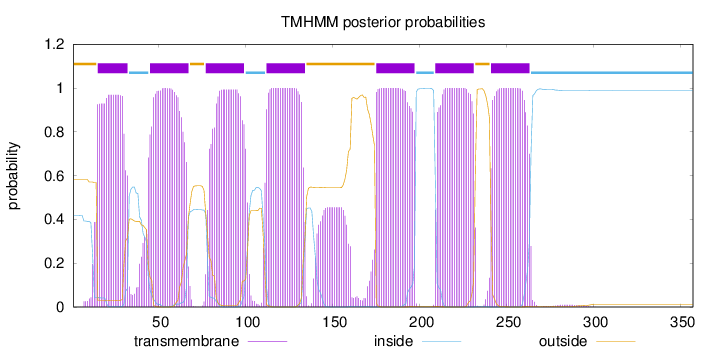
Topology
Length:
357
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
162.2285
Exp number, first 60 AAs:
35.4025
Total prob of N-in:
0.41776
POSSIBLE N-term signal
sequence
outside
1 - 14
TMhelix
15 - 32
inside
33 - 44
TMhelix
45 - 67
outside
68 - 76
TMhelix
77 - 99
inside
100 - 111
TMhelix
112 - 134
outside
135 - 174
TMhelix
175 - 197
inside
198 - 208
TMhelix
209 - 231
outside
232 - 240
TMhelix
241 - 263
inside
264 - 357
Population Genetic Test Statistics
Pi
306.814281
Theta
183.164296
Tajima's D
1.919526
CLR
0.005114
CSRT
0.866656667166642
Interpretation
Uncertain
