Gene
KWMTBOMO10930
Pre Gene Modal
BGIBMGA008342
Annotation
PREDICTED:_NEDD8-activating_enzyme_E1_regulatory_subunit_[Papilio_machaon]
Full name
NEDD8-activating enzyme E1 regulatory subunit
Alternative Name
APP-BP1
Amyloid protein-binding protein 1
Amyloid protein-binding protein 1
Location in the cell
Cytoplasmic Reliability : 3.488
Sequence
CDS
ATGGCGTCTCCATCTCCTAAATCACCAGAACAAAGCGAGAAGAATAAACAGTACGACCGGCAACTTCGTTTGTGGGGTGATCACGGACAAAGTGCTCTTGAAAGCGGACACATATGTTTAATTAATGCTACTGCCTTGGGTACTGAAATATTGAAATCGATAGTTCTTCCAGGAATTGGTGCAATTACTATAGTTGATGATTGCACAGTTAGTGATGAAGATATAGGCAGTAATTTCTTTTTGGAAATGTCTAGCAAAGGTCTTAACAGAGGATCCGAAGCGTTAAGGCTTGTTTTAGAATTAAATCCAGCAGTACAAGGTCATGCAGTCCAAGAACCACCTGACCAGATACTTGATGAGAACCCAGACTTTTTTAAATCATTCAGTTTAGTGATAGCTACTACAATAAGTGAAAAAGTAACGCAAAGTTTATCTCAACACCTGTGGGATATAAATGTTCCTTTTATACTTTGCCAATCAGTTGGATTCTTAGGGTCTTTAAGAATTCAGATTCGGGAACATGCTGTTACTGAAACTCATCCTGATAATGAACAAACAGATCTGCGCCTAGATGAACCATTTATGTCATTGTTAGAACATCTGGAAAGTATTGATATTGATGCTCTTGATTTGAAAGATCATGGTCACATTCCTTGGATTATAATTCTTTATAAAGCTGTTCAAAAGTGGAAAATTAACAATAACAATAAATGGCCTGTAACGCGAAATGATAAAGAAGATATAAAAAATATAATAAGGGATTTCATTAGAAAAGATGAAAATGGAATACCAATTAATGAAGAGAATTTTGAGGAAGCAATAAGAGCTGTTAACACTGCTTTGATGCCTACATTTTTGCCTAAGAATATCTCAAAACTTATGTATAATAGTGCTGCCACAAATTTAACTAAGGATAGTTCATCATTTTGGATTATGTGCTCTGCCTTAAGAGCATTTGTTGAGGCTGAAGGGAAGGGAAAACTTCCAGTGAAAGGAATCTTACCTGATATGACGGCCTCTACAGAACATTATGTAAAACTTCAAAACATTTACAAGATGCAAGCTGCAAATGATGCAGAAATAGTCTATAGAAAAGTTCGACAGATAGTCACTCAATTGCATTGTGATAGTATTAGTGAGGCAGATGTTAAACTATTCTGCAGACATGCTCATGATTTATATTTGGTGAAGGGGTCTTGTGTGGCAACAGAACTTCAACTGGGAAATTCTGTTGCATCCTATATAGCCACATATTTAGAGGAGCCAGATGTCATGATGGTTCACTATATTATGCTTCGGGCAGCTGGTATGTTTAGATCAGAGCACTGCCGAGCTCCCGGTGATTGGGAACCAGAAGGTGACATTGCTAAGCTAAAAACTTGTGTAAATAAACTTTTAGGTGATATATCTTGTTCTCCATTTCCTAAAGATGATCATATCCACGAAATGTGTCGCTATGGGGGAGCCGAGATACATAGTGTGTCTGCCTTTTTAGGTGGATGTGTTGCACATGAAGCCATTAAAATTATAACCAAGCAATATAAACCTGTAAATAATACATTTATTTATGATGGAGCTTCTACAAATACGGCAACATTTGAATTCTAA
Protein
MASPSPKSPEQSEKNKQYDRQLRLWGDHGQSALESGHICLINATALGTEILKSIVLPGIGAITIVDDCTVSDEDIGSNFFLEMSSKGLNRGSEALRLVLELNPAVQGHAVQEPPDQILDENPDFFKSFSLVIATTISEKVTQSLSQHLWDINVPFILCQSVGFLGSLRIQIREHAVTETHPDNEQTDLRLDEPFMSLLEHLESIDIDALDLKDHGHIPWIIILYKAVQKWKINNNNKWPVTRNDKEDIKNIIRDFIRKDENGIPINEENFEEAIRAVNTALMPTFLPKNISKLMYNSAATNLTKDSSSFWIMCSALRAFVEAEGKGKLPVKGILPDMTASTEHYVKLQNIYKMQAANDAEIVYRKVRQIVTQLHCDSISEADVKLFCRHAHDLYLVKGSCVATELQLGNSVASYIATYLEEPDVMMVHYIMLRAAGMFRSEHCRAPGDWEPEGDIAKLKTCVNKLLGDISCSPFPKDDHIHEMCRYGGAEIHSVSAFLGGCVAHEAIKIITKQYKPVNNTFIYDGASTNTATFEF
Summary
Description
Regulatory subunit of the dimeric UBA3-NAE1 E1 enzyme. E1 activates NEDD8 by first adenylating its C-terminal glycine residue with ATP, thereafter linking this residue to the side chain of the catalytic cysteine, yielding a NEDD8-UBA3 thioester and free AMP. E1 finally transfers NEDD8 to the catalytic cysteine of UBE2M.
Regulatory subunit of the dimeric uba3-nae1 E1 enzyme. E1 activates nedd8 by first adenylating its C-terminal glycine residue with ATP, thereafter linking this residue to the side chain of the catalytic cysteine, yielding a nedd8-uba3 thioester and free AMP. E1 finally transfers nedd8 to the catalytic cysteine of ube2m (By similarity).
Regulatory subunit of the dimeric uba3-nae1 E1 enzyme. E1 activates nedd8 by first adenylating its C-terminal glycine residue with ATP, thereafter linking this residue to the side chain of the catalytic cysteine, yielding a nedd8-uba3 thioester and free AMP. E1 finally transfers nedd8 to the catalytic cysteine of ube2m (By similarity).
Subunit
Heterodimer of uba3 and nae1. The complex binds nedd8 and ube2m (By similarity).
Similarity
Belongs to the ubiquitin-activating E1 family. ULA1 subfamily.
Keywords
Ubl conjugation pathway
Alternative splicing
Complete proteome
Reference proteome
Feature
chain NEDD8-activating enzyme E1 regulatory subunit
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9JFP5
A0A2A4K5E5
A0A2H1WS32
A0A2W1C107
A0A194RQD7
A0A194PGP9
+ More
A0A212FCA1 A0A0L7KU08 A0A026WUW0 A0A1B6EM87 A0A2J7REN5 E9IAZ2 A0A067RIK4 K7IP66 A0A154P9P5 E2AY86 A0A1B6BZM3 A0A0C9QMW4 A0A232FA34 A0A151XDN3 E2B689 A0A151IL80 A0A0M8ZUW6 A0A151IYU0 A0A146LYI4 A0A158P3I5 E0W0X0 A0A151JXD6 A0A0L7RDR6 A0A0V0G5L6 A0A224XN02 A0A2A3E8Y8 A0A0P4VKL2 T1I595 R4G5L8 A0A1S3CZ93 J9K171 A0A131Z5N2 L7MAB5 D6WGG4 A0A2S2PFJ2 A0A224YJJ3 A0A131XH62 A0A131XSV5 A0A3P9JD45 A0A0P4VXZ9 A0A147BSN8 A0A3P9L0Q9 H2LLC6 A0A1E1X2X7 A0A3B5B099 A0A182GEJ2 A0A1W4ZWL0 F4WB76 A0A3Q3R0L6 A0A3Q3KPK7 A0A1E1XSX6 G3MPB9 A0A1W5A5M5 A0A336MK03 A0A3B3DS61 A0A3B4WD32 A0A336LIK4 A0A023GN81 A0A3Q2ZBP2 A0A2K6VB61 J7M297 A0A3S2PRV1 A0A3Q1I6M6 A0A2U9BBW2 A0A1Q3FA90 B0X7Y7 A0A084WKK0 A0A3Q1C1Y7 A0A2M4CYD2 Q7Q726 Q6NTW6 A0A1L8GEL9 A0A1J1IPP2 Q170Z9 A0A182LJF3 A0A3P8TPC3 Q7SXP2 B0XB59 A0A182MK24 A0A182XUF7 A0A3Q2PTC9 A0A1S3KTD2 A0A2R5LCJ3 F1QR83 I3J4P7 B7ZV51 B0BME5 H2V4P7 A0A3P9AZQ1 A0A3Q2WBR0 Q16N42 A0A3B4F5T3 A0A3P8ULS9
A0A212FCA1 A0A0L7KU08 A0A026WUW0 A0A1B6EM87 A0A2J7REN5 E9IAZ2 A0A067RIK4 K7IP66 A0A154P9P5 E2AY86 A0A1B6BZM3 A0A0C9QMW4 A0A232FA34 A0A151XDN3 E2B689 A0A151IL80 A0A0M8ZUW6 A0A151IYU0 A0A146LYI4 A0A158P3I5 E0W0X0 A0A151JXD6 A0A0L7RDR6 A0A0V0G5L6 A0A224XN02 A0A2A3E8Y8 A0A0P4VKL2 T1I595 R4G5L8 A0A1S3CZ93 J9K171 A0A131Z5N2 L7MAB5 D6WGG4 A0A2S2PFJ2 A0A224YJJ3 A0A131XH62 A0A131XSV5 A0A3P9JD45 A0A0P4VXZ9 A0A147BSN8 A0A3P9L0Q9 H2LLC6 A0A1E1X2X7 A0A3B5B099 A0A182GEJ2 A0A1W4ZWL0 F4WB76 A0A3Q3R0L6 A0A3Q3KPK7 A0A1E1XSX6 G3MPB9 A0A1W5A5M5 A0A336MK03 A0A3B3DS61 A0A3B4WD32 A0A336LIK4 A0A023GN81 A0A3Q2ZBP2 A0A2K6VB61 J7M297 A0A3S2PRV1 A0A3Q1I6M6 A0A2U9BBW2 A0A1Q3FA90 B0X7Y7 A0A084WKK0 A0A3Q1C1Y7 A0A2M4CYD2 Q7Q726 Q6NTW6 A0A1L8GEL9 A0A1J1IPP2 Q170Z9 A0A182LJF3 A0A3P8TPC3 Q7SXP2 B0XB59 A0A182MK24 A0A182XUF7 A0A3Q2PTC9 A0A1S3KTD2 A0A2R5LCJ3 F1QR83 I3J4P7 B7ZV51 B0BME5 H2V4P7 A0A3P9AZQ1 A0A3Q2WBR0 Q16N42 A0A3B4F5T3 A0A3P8ULS9
Pubmed
19121390
28756777
26354079
22118469
26227816
24508170
+ More
30249741 21282665 24845553 20075255 20798317 28648823 26823975 21347285 20566863 27129103 26830274 25576852 18362917 19820115 28797301 28049606 17554307 29652888 28503490 26483478 21719571 29209593 22216098 29451363 20920257 24438588 12364791 14747013 17210077 27762356 17510324 20966253 23594743 25186727 20431018 21551351 24487278
30249741 21282665 24845553 20075255 20798317 28648823 26823975 21347285 20566863 27129103 26830274 25576852 18362917 19820115 28797301 28049606 17554307 29652888 28503490 26483478 21719571 29209593 22216098 29451363 20920257 24438588 12364791 14747013 17210077 27762356 17510324 20966253 23594743 25186727 20431018 21551351 24487278
EMBL
BABH01001984
NWSH01000165
PCG78902.1
ODYU01010245
SOQ55224.1
KZ149920
+ More
PZC77763.1 KQ459765 KPJ20078.1 KQ459604 KPI92561.1 AGBW02009208 OWR51362.1 JTDY01005869 KOB66534.1 KK107088 QOIP01000008 EZA59802.1 RLU19506.1 GECZ01030746 JAS39023.1 NEVH01004959 PNF39299.1 GL762091 EFZ22238.1 KK852482 KDR22868.1 KQ434842 KZC08114.1 GL443762 EFN61621.1 GEDC01030586 JAS06712.1 GBYB01004869 GBYB01004870 JAG74636.1 JAG74637.1 NNAY01000633 OXU27293.1 KQ982268 KYQ58463.1 GL445930 EFN88782.1 KQ977133 KYN05489.1 KQ435840 KOX71457.1 KQ980740 KYN13719.1 GDHC01006400 JAQ12229.1 ADTU01008083 DS235862 EEB19276.1 KQ981604 KYN39717.1 KQ414612 KOC69132.1 GECL01002764 JAP03360.1 GFTR01007025 JAW09401.1 KZ288344 PBC27666.1 GDKW01000968 JAI55627.1 ACPB03000705 GAHY01000420 JAA77090.1 ABLF02040187 GEDV01002269 JAP86288.1 GACK01004956 JAA60078.1 KQ971321 EFA00552.1 GGMR01015590 MBY28209.1 GFPF01005979 MAA17125.1 GEFH01003153 JAP65428.1 GEFM01006488 JAP69308.1 GDRN01104620 JAI57881.1 GEGO01001620 JAR93784.1 GFAC01005563 JAT93625.1 JXUM01242700 KQ635604 KXJ62460.1 GL888058 EGI68549.1 GFAA01001042 JAU02393.1 JO843720 AEO35337.1 UFQS01000673 UFQT01000673 SSX06031.1 SSX26388.1 UFQS01004372 UFQT01004372 SSX16351.1 SSX35668.1 GBBM01000815 JAC34603.1 AB665323 BAM36364.1 CM012439 RVE74384.1 CP026247 AWP01443.1 GFDL01010606 JAV24439.1 DS232468 EDS42206.1 ATLV01024123 KE525349 KFB50744.1 GGFL01006097 MBW70275.1 AAAB01008960 EAA11771.2 BC068838 CM004473 OCT82328.1 CVRI01000057 CRL02213.1 CH477462 EAT40545.1 BC050171 BC055513 BC151887 DS232614 EDS44112.1 AXCM01007885 GGLE01003117 MBY07243.1 BX511136 AERX01013865 BC171459 AAI71459.1 AAMC01025501 BC158396 AAI58397.1 CH477841 EAT35744.1
PZC77763.1 KQ459765 KPJ20078.1 KQ459604 KPI92561.1 AGBW02009208 OWR51362.1 JTDY01005869 KOB66534.1 KK107088 QOIP01000008 EZA59802.1 RLU19506.1 GECZ01030746 JAS39023.1 NEVH01004959 PNF39299.1 GL762091 EFZ22238.1 KK852482 KDR22868.1 KQ434842 KZC08114.1 GL443762 EFN61621.1 GEDC01030586 JAS06712.1 GBYB01004869 GBYB01004870 JAG74636.1 JAG74637.1 NNAY01000633 OXU27293.1 KQ982268 KYQ58463.1 GL445930 EFN88782.1 KQ977133 KYN05489.1 KQ435840 KOX71457.1 KQ980740 KYN13719.1 GDHC01006400 JAQ12229.1 ADTU01008083 DS235862 EEB19276.1 KQ981604 KYN39717.1 KQ414612 KOC69132.1 GECL01002764 JAP03360.1 GFTR01007025 JAW09401.1 KZ288344 PBC27666.1 GDKW01000968 JAI55627.1 ACPB03000705 GAHY01000420 JAA77090.1 ABLF02040187 GEDV01002269 JAP86288.1 GACK01004956 JAA60078.1 KQ971321 EFA00552.1 GGMR01015590 MBY28209.1 GFPF01005979 MAA17125.1 GEFH01003153 JAP65428.1 GEFM01006488 JAP69308.1 GDRN01104620 JAI57881.1 GEGO01001620 JAR93784.1 GFAC01005563 JAT93625.1 JXUM01242700 KQ635604 KXJ62460.1 GL888058 EGI68549.1 GFAA01001042 JAU02393.1 JO843720 AEO35337.1 UFQS01000673 UFQT01000673 SSX06031.1 SSX26388.1 UFQS01004372 UFQT01004372 SSX16351.1 SSX35668.1 GBBM01000815 JAC34603.1 AB665323 BAM36364.1 CM012439 RVE74384.1 CP026247 AWP01443.1 GFDL01010606 JAV24439.1 DS232468 EDS42206.1 ATLV01024123 KE525349 KFB50744.1 GGFL01006097 MBW70275.1 AAAB01008960 EAA11771.2 BC068838 CM004473 OCT82328.1 CVRI01000057 CRL02213.1 CH477462 EAT40545.1 BC050171 BC055513 BC151887 DS232614 EDS44112.1 AXCM01007885 GGLE01003117 MBY07243.1 BX511136 AERX01013865 BC171459 AAI71459.1 AAMC01025501 BC158396 AAI58397.1 CH477841 EAT35744.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
+ More
UP000053097 UP000279307 UP000235965 UP000027135 UP000002358 UP000076502 UP000000311 UP000215335 UP000075809 UP000008237 UP000078542 UP000053105 UP000078492 UP000005205 UP000009046 UP000078541 UP000053825 UP000242457 UP000015103 UP000079169 UP000007819 UP000007266 UP000265200 UP000265180 UP000001038 UP000261400 UP000069940 UP000249989 UP000192224 UP000007755 UP000261600 UP000261640 UP000261560 UP000261360 UP000264800 UP000265040 UP000246464 UP000002320 UP000030765 UP000257160 UP000007062 UP000186698 UP000183832 UP000008820 UP000075882 UP000265080 UP000000437 UP000075883 UP000076407 UP000265000 UP000087266 UP000005207 UP000008143 UP000005226 UP000265160 UP000264840 UP000261460 UP000265120
UP000053097 UP000279307 UP000235965 UP000027135 UP000002358 UP000076502 UP000000311 UP000215335 UP000075809 UP000008237 UP000078542 UP000053105 UP000078492 UP000005205 UP000009046 UP000078541 UP000053825 UP000242457 UP000015103 UP000079169 UP000007819 UP000007266 UP000265200 UP000265180 UP000001038 UP000261400 UP000069940 UP000249989 UP000192224 UP000007755 UP000261600 UP000261640 UP000261560 UP000261360 UP000264800 UP000265040 UP000246464 UP000002320 UP000030765 UP000257160 UP000007062 UP000186698 UP000183832 UP000008820 UP000075882 UP000265080 UP000000437 UP000075883 UP000076407 UP000265000 UP000087266 UP000005207 UP000008143 UP000005226 UP000265160 UP000264840 UP000261460 UP000265120
PRIDE
Pfam
PF00899 ThiF
SUPFAM
SSF69572
SSF69572
ProteinModelPortal
H9JFP5
A0A2A4K5E5
A0A2H1WS32
A0A2W1C107
A0A194RQD7
A0A194PGP9
+ More
A0A212FCA1 A0A0L7KU08 A0A026WUW0 A0A1B6EM87 A0A2J7REN5 E9IAZ2 A0A067RIK4 K7IP66 A0A154P9P5 E2AY86 A0A1B6BZM3 A0A0C9QMW4 A0A232FA34 A0A151XDN3 E2B689 A0A151IL80 A0A0M8ZUW6 A0A151IYU0 A0A146LYI4 A0A158P3I5 E0W0X0 A0A151JXD6 A0A0L7RDR6 A0A0V0G5L6 A0A224XN02 A0A2A3E8Y8 A0A0P4VKL2 T1I595 R4G5L8 A0A1S3CZ93 J9K171 A0A131Z5N2 L7MAB5 D6WGG4 A0A2S2PFJ2 A0A224YJJ3 A0A131XH62 A0A131XSV5 A0A3P9JD45 A0A0P4VXZ9 A0A147BSN8 A0A3P9L0Q9 H2LLC6 A0A1E1X2X7 A0A3B5B099 A0A182GEJ2 A0A1W4ZWL0 F4WB76 A0A3Q3R0L6 A0A3Q3KPK7 A0A1E1XSX6 G3MPB9 A0A1W5A5M5 A0A336MK03 A0A3B3DS61 A0A3B4WD32 A0A336LIK4 A0A023GN81 A0A3Q2ZBP2 A0A2K6VB61 J7M297 A0A3S2PRV1 A0A3Q1I6M6 A0A2U9BBW2 A0A1Q3FA90 B0X7Y7 A0A084WKK0 A0A3Q1C1Y7 A0A2M4CYD2 Q7Q726 Q6NTW6 A0A1L8GEL9 A0A1J1IPP2 Q170Z9 A0A182LJF3 A0A3P8TPC3 Q7SXP2 B0XB59 A0A182MK24 A0A182XUF7 A0A3Q2PTC9 A0A1S3KTD2 A0A2R5LCJ3 F1QR83 I3J4P7 B7ZV51 B0BME5 H2V4P7 A0A3P9AZQ1 A0A3Q2WBR0 Q16N42 A0A3B4F5T3 A0A3P8ULS9
A0A212FCA1 A0A0L7KU08 A0A026WUW0 A0A1B6EM87 A0A2J7REN5 E9IAZ2 A0A067RIK4 K7IP66 A0A154P9P5 E2AY86 A0A1B6BZM3 A0A0C9QMW4 A0A232FA34 A0A151XDN3 E2B689 A0A151IL80 A0A0M8ZUW6 A0A151IYU0 A0A146LYI4 A0A158P3I5 E0W0X0 A0A151JXD6 A0A0L7RDR6 A0A0V0G5L6 A0A224XN02 A0A2A3E8Y8 A0A0P4VKL2 T1I595 R4G5L8 A0A1S3CZ93 J9K171 A0A131Z5N2 L7MAB5 D6WGG4 A0A2S2PFJ2 A0A224YJJ3 A0A131XH62 A0A131XSV5 A0A3P9JD45 A0A0P4VXZ9 A0A147BSN8 A0A3P9L0Q9 H2LLC6 A0A1E1X2X7 A0A3B5B099 A0A182GEJ2 A0A1W4ZWL0 F4WB76 A0A3Q3R0L6 A0A3Q3KPK7 A0A1E1XSX6 G3MPB9 A0A1W5A5M5 A0A336MK03 A0A3B3DS61 A0A3B4WD32 A0A336LIK4 A0A023GN81 A0A3Q2ZBP2 A0A2K6VB61 J7M297 A0A3S2PRV1 A0A3Q1I6M6 A0A2U9BBW2 A0A1Q3FA90 B0X7Y7 A0A084WKK0 A0A3Q1C1Y7 A0A2M4CYD2 Q7Q726 Q6NTW6 A0A1L8GEL9 A0A1J1IPP2 Q170Z9 A0A182LJF3 A0A3P8TPC3 Q7SXP2 B0XB59 A0A182MK24 A0A182XUF7 A0A3Q2PTC9 A0A1S3KTD2 A0A2R5LCJ3 F1QR83 I3J4P7 B7ZV51 B0BME5 H2V4P7 A0A3P9AZQ1 A0A3Q2WBR0 Q16N42 A0A3B4F5T3 A0A3P8ULS9
PDB
1YOV
E-value=5.8486e-144,
Score=1311
Ontologies
GO
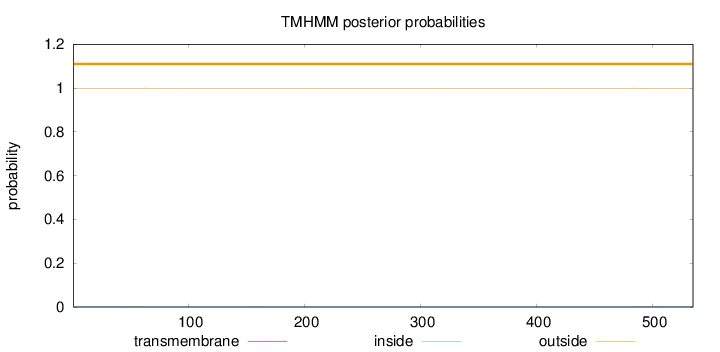
Topology
Length:
535
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02674
Exp number, first 60 AAs:
0.00433
Total prob of N-in:
0.00070
outside
1 - 535
Population Genetic Test Statistics
Pi
118.877245
Theta
200.533811
Tajima's D
-1.526547
CLR
2983.017664
CSRT
0.0607469626518674
Interpretation
Uncertain
