Gene
KWMTBOMO10923
Pre Gene Modal
BGIBMGA008493
Annotation
putative_small_nuclear_ribonucleoprotein?_core_[Danaus_plexippus]
Location in the cell
Cytoplasmic Reliability : 2.048
Sequence
CDS
ATGAGTAATGAAGGCAATAAAACGACTTCGGAAGCACCTCAGGTGGTGGAGGAAGAAGGAAAGGCAAAACTTAGAAAATGGTTAAATATGAAATTTAGAATTGAAATGACCGATGGCAGAGTTCTGATAGGAGTGTTCCTGTGTACAGATAGAGATGCAAATGTAATTTTAGGTGCTTGCTCAGAGTACTTGAAAAATAATGAGGGAGAAACTGAAGAACCCAGAGTATTAGGTCTAGTCATGGTTCCTGGAAGACACATTGTGTCAATACAAATTGATGATACAACACCATCTCAAACATACTATTATGATGAATATCTAATTAAATAA
Protein
MSNEGNKTTSEAPQVVEEEGKAKLRKWLNMKFRIEMTDGRVLIGVFLCTDRDANVILGACSEYLKNNEGETEEPRVLGLVMVPGRHIVSIQIDDTTPSQTYYYDEYLIK
Summary
Uniprot
H9JG46
A0A212FCB4
S4PMS9
A0A2H1WKP2
A0A2A4JS06
A0A194PHP6
+ More
A0A194RRS0 A0A067RL84 R4UW31 A0A0C9S9Z9 A0A194ANC0 K7INW7 A0A232FLP9 U4UG11 N6SYY1 A0A210PXR8 A0A131YR78 A0A224YST4 A0A131XQZ5 A0A195EU77 F4W485 A0A158NFL2 A0A195E4R7 A0A195B3H4 A0A1B6LDJ9 A0A146LJY8 A0A0P4VQ73 A0A146LUS3 A0A0A9Y3E5 A0A151X1M5 A0A195CKE6 A0A026VVZ2 A0A087T6F7 A0A1E1WW84 A0A2R5LES3 A0A147BF49 A0A0T6B2R1 A0A1B6JYY9 A0A1S3I4G4 A0A224XNN6 D1ZZY7 A0A0P4VL70 R4G2Z0 A0A069DPA1 T1J0V0 A0A1B6M5T1 A0A0L7RII3 A0A2P6KL38 A0A0B6ZEJ4 A0A2A3ECX9 V9IC93 A0A088ARK8 B4MZZ5 A0A154P616 A0A0L8GUA9 T1FZF6 A0A1B6CL54 A0A1Y1LNE1 A0A0M9A7S8 A0A0P4XG69 A0A0N8BF97 A0A1Y1LNP4 A0A164Z295 A0A0P5FH00 A0A3B4CRS2 B4GSB4 Q29KG1 A0A2H8TLA3 A0A2S2NC48 A0A3S1BQ87 C4WXE2 A0A0K8UGC6 B4M9I3 A0A3B5QXW0 A0A3B5KTG5 A0A1S3DR92 A0A1L8DGG3 A0A1L8DG98 A0A2P2IA21 A0A2R7X890 A0A1W4UTH2 A0A2S2QJH9 A0A3B0JND8 T1HHZ1 B4Q8M7 Q8IPZ7 A0A1S3D5N1 B3NAM4 A0A3B5K1M6 V4BNL3 Q4V620 A0A3B1JU85 B0XE29 A0A3P8YUD7
A0A194RRS0 A0A067RL84 R4UW31 A0A0C9S9Z9 A0A194ANC0 K7INW7 A0A232FLP9 U4UG11 N6SYY1 A0A210PXR8 A0A131YR78 A0A224YST4 A0A131XQZ5 A0A195EU77 F4W485 A0A158NFL2 A0A195E4R7 A0A195B3H4 A0A1B6LDJ9 A0A146LJY8 A0A0P4VQ73 A0A146LUS3 A0A0A9Y3E5 A0A151X1M5 A0A195CKE6 A0A026VVZ2 A0A087T6F7 A0A1E1WW84 A0A2R5LES3 A0A147BF49 A0A0T6B2R1 A0A1B6JYY9 A0A1S3I4G4 A0A224XNN6 D1ZZY7 A0A0P4VL70 R4G2Z0 A0A069DPA1 T1J0V0 A0A1B6M5T1 A0A0L7RII3 A0A2P6KL38 A0A0B6ZEJ4 A0A2A3ECX9 V9IC93 A0A088ARK8 B4MZZ5 A0A154P616 A0A0L8GUA9 T1FZF6 A0A1B6CL54 A0A1Y1LNE1 A0A0M9A7S8 A0A0P4XG69 A0A0N8BF97 A0A1Y1LNP4 A0A164Z295 A0A0P5FH00 A0A3B4CRS2 B4GSB4 Q29KG1 A0A2H8TLA3 A0A2S2NC48 A0A3S1BQ87 C4WXE2 A0A0K8UGC6 B4M9I3 A0A3B5QXW0 A0A3B5KTG5 A0A1S3DR92 A0A1L8DGG3 A0A1L8DG98 A0A2P2IA21 A0A2R7X890 A0A1W4UTH2 A0A2S2QJH9 A0A3B0JND8 T1HHZ1 B4Q8M7 Q8IPZ7 A0A1S3D5N1 B3NAM4 A0A3B5K1M6 V4BNL3 Q4V620 A0A3B1JU85 B0XE29 A0A3P8YUD7
Pubmed
19121390
22118469
23622113
26354079
24845553
26131772
+ More
20075255 28648823 23537049 28812685 26830274 28797301 28049606 21719571 21347285 26823975 25401762 24508170 30249741 28503490 29652888 18362917 19820115 27129103 26334808 17994087 23254933 28004739 15632085 23542700 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21551351 25329095 25069045
20075255 28648823 23537049 28812685 26830274 28797301 28049606 21719571 21347285 26823975 25401762 24508170 30249741 28503490 29652888 18362917 19820115 27129103 26334808 17994087 23254933 28004739 15632085 23542700 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21551351 25329095 25069045
EMBL
BABH01001974
BABH01001975
AGBW02009208
OWR51358.1
GAIX01000016
JAA92544.1
+ More
ODYU01008976 SOQ53024.1 NWSH01000789 PCG74190.1 KQ459604 KPI92568.1 KQ459765 KPJ20084.1 KK852559 KDR21370.1 KC571937 AGM32436.1 GBZX01002888 JAG89852.1 GELH01000550 GELH01000549 JAS03723.1 NNAY01000039 OXU31676.1 KB632308 ERL91962.1 APGK01051610 KB741197 ENN72979.1 NEDP02005415 OWF41264.1 GEDV01006798 JAP81759.1 GFPF01009490 MAA20636.1 GEFH01000081 JAP68500.1 KQ981979 KYN31457.1 GL887491 EGI71081.1 ADTU01001535 KQ979609 KYN20180.1 KQ976625 KYM79043.1 GEBQ01018206 JAT21771.1 GDHC01010216 JAQ08413.1 GDRN01108840 JAI57159.1 GDHC01008157 JAQ10472.1 GBHO01017996 GBHO01017994 GBRD01013963 GDHC01001078 JAG25608.1 JAG25610.1 JAG51863.1 JAQ17551.1 KQ982585 KYQ54311.1 KQ977642 KYN00927.1 KK107796 QOIP01000013 EZA47656.1 RLU15576.1 KK113646 KFM60696.1 GFAC01007903 JAT91285.1 GGLE01003895 MBY08021.1 GEGO01006011 JAR89393.1 LJIG01016057 KRT81743.1 GECU01003316 JAT04391.1 GFTR01002331 JAW14095.1 KQ971338 EFA02441.1 GDKW01003264 JAI53331.1 ACPB03011258 ACPB03011259 GAHY01002161 JAA75349.1 GBGD01003442 JAC85447.1 AFFK01020674 GEBQ01008709 JAT31268.1 KQ414584 KOC70639.1 MWRG01009489 PRD27048.1 HACG01019416 CEK66281.1 KZ288282 PBC29587.1 JR039434 AEY58700.1 CH963920 EDW77930.1 KQ434825 KZC07375.1 KQ420471 KOF80185.1 AMQM01001337 KB097495 ESN96169.1 GEDC01023146 JAS14152.1 GEZM01052525 GEZM01052524 JAV74461.1 KQ435726 KOX78216.1 GDIP01244791 JAI78610.1 GDIQ01183483 JAK68242.1 GEZM01052526 GEZM01052523 JAV74458.1 LRGB01000818 KZS15864.1 GDIQ01258245 JAJ93479.1 CH479189 EDW25661.1 CH379061 EAL33214.2 GFXV01003131 GFXV01003786 MBW14936.1 MBW15591.1 GGMR01001903 MBY14522.1 RQTK01000072 RUS88865.1 ABLF02029770 AK342591 BAH72562.1 GDHF01026691 JAI25623.1 CH940654 EDW57859.1 GFDF01008563 JAV05521.1 GFDF01008605 JAV05479.1 IACF01005279 LAB70865.1 KK858134 PTY27650.1 GGMS01008716 MBY77919.1 OUUW01000006 SPP82393.1 ACPB03018543 CM000361 CM002910 EDX03544.1 KMY87772.1 AE014134 BT044300 KX531912 AAN10396.1 ACH92365.1 AGB92508.1 ANY27722.1 CH954177 EDV57547.1 KB202408 ESO90454.1 BT022486 AAY54902.1 DS232799 EDS45801.1
ODYU01008976 SOQ53024.1 NWSH01000789 PCG74190.1 KQ459604 KPI92568.1 KQ459765 KPJ20084.1 KK852559 KDR21370.1 KC571937 AGM32436.1 GBZX01002888 JAG89852.1 GELH01000550 GELH01000549 JAS03723.1 NNAY01000039 OXU31676.1 KB632308 ERL91962.1 APGK01051610 KB741197 ENN72979.1 NEDP02005415 OWF41264.1 GEDV01006798 JAP81759.1 GFPF01009490 MAA20636.1 GEFH01000081 JAP68500.1 KQ981979 KYN31457.1 GL887491 EGI71081.1 ADTU01001535 KQ979609 KYN20180.1 KQ976625 KYM79043.1 GEBQ01018206 JAT21771.1 GDHC01010216 JAQ08413.1 GDRN01108840 JAI57159.1 GDHC01008157 JAQ10472.1 GBHO01017996 GBHO01017994 GBRD01013963 GDHC01001078 JAG25608.1 JAG25610.1 JAG51863.1 JAQ17551.1 KQ982585 KYQ54311.1 KQ977642 KYN00927.1 KK107796 QOIP01000013 EZA47656.1 RLU15576.1 KK113646 KFM60696.1 GFAC01007903 JAT91285.1 GGLE01003895 MBY08021.1 GEGO01006011 JAR89393.1 LJIG01016057 KRT81743.1 GECU01003316 JAT04391.1 GFTR01002331 JAW14095.1 KQ971338 EFA02441.1 GDKW01003264 JAI53331.1 ACPB03011258 ACPB03011259 GAHY01002161 JAA75349.1 GBGD01003442 JAC85447.1 AFFK01020674 GEBQ01008709 JAT31268.1 KQ414584 KOC70639.1 MWRG01009489 PRD27048.1 HACG01019416 CEK66281.1 KZ288282 PBC29587.1 JR039434 AEY58700.1 CH963920 EDW77930.1 KQ434825 KZC07375.1 KQ420471 KOF80185.1 AMQM01001337 KB097495 ESN96169.1 GEDC01023146 JAS14152.1 GEZM01052525 GEZM01052524 JAV74461.1 KQ435726 KOX78216.1 GDIP01244791 JAI78610.1 GDIQ01183483 JAK68242.1 GEZM01052526 GEZM01052523 JAV74458.1 LRGB01000818 KZS15864.1 GDIQ01258245 JAJ93479.1 CH479189 EDW25661.1 CH379061 EAL33214.2 GFXV01003131 GFXV01003786 MBW14936.1 MBW15591.1 GGMR01001903 MBY14522.1 RQTK01000072 RUS88865.1 ABLF02029770 AK342591 BAH72562.1 GDHF01026691 JAI25623.1 CH940654 EDW57859.1 GFDF01008563 JAV05521.1 GFDF01008605 JAV05479.1 IACF01005279 LAB70865.1 KK858134 PTY27650.1 GGMS01008716 MBY77919.1 OUUW01000006 SPP82393.1 ACPB03018543 CM000361 CM002910 EDX03544.1 KMY87772.1 AE014134 BT044300 KX531912 AAN10396.1 ACH92365.1 AGB92508.1 ANY27722.1 CH954177 EDV57547.1 KB202408 ESO90454.1 BT022486 AAY54902.1 DS232799 EDS45801.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053268
UP000053240
UP000027135
+ More
UP000002358 UP000215335 UP000030742 UP000019118 UP000242188 UP000078541 UP000007755 UP000005205 UP000078492 UP000078540 UP000075809 UP000078542 UP000053097 UP000279307 UP000054359 UP000085678 UP000007266 UP000015103 UP000053825 UP000242457 UP000005203 UP000007798 UP000076502 UP000053454 UP000015101 UP000053105 UP000076858 UP000261440 UP000008744 UP000001819 UP000271974 UP000007819 UP000008792 UP000002852 UP000261380 UP000079169 UP000192221 UP000268350 UP000000304 UP000000803 UP000008711 UP000005226 UP000030746 UP000018467 UP000002320 UP000265140
UP000002358 UP000215335 UP000030742 UP000019118 UP000242188 UP000078541 UP000007755 UP000005205 UP000078492 UP000078540 UP000075809 UP000078542 UP000053097 UP000279307 UP000054359 UP000085678 UP000007266 UP000015103 UP000053825 UP000242457 UP000005203 UP000007798 UP000076502 UP000053454 UP000015101 UP000053105 UP000076858 UP000261440 UP000008744 UP000001819 UP000271974 UP000007819 UP000008792 UP000002852 UP000261380 UP000079169 UP000192221 UP000268350 UP000000304 UP000000803 UP000008711 UP000005226 UP000030746 UP000018467 UP000002320 UP000265140
Pfam
PF01423 LSM
SUPFAM
SSF50182
SSF50182
CDD
ProteinModelPortal
H9JG46
A0A212FCB4
S4PMS9
A0A2H1WKP2
A0A2A4JS06
A0A194PHP6
+ More
A0A194RRS0 A0A067RL84 R4UW31 A0A0C9S9Z9 A0A194ANC0 K7INW7 A0A232FLP9 U4UG11 N6SYY1 A0A210PXR8 A0A131YR78 A0A224YST4 A0A131XQZ5 A0A195EU77 F4W485 A0A158NFL2 A0A195E4R7 A0A195B3H4 A0A1B6LDJ9 A0A146LJY8 A0A0P4VQ73 A0A146LUS3 A0A0A9Y3E5 A0A151X1M5 A0A195CKE6 A0A026VVZ2 A0A087T6F7 A0A1E1WW84 A0A2R5LES3 A0A147BF49 A0A0T6B2R1 A0A1B6JYY9 A0A1S3I4G4 A0A224XNN6 D1ZZY7 A0A0P4VL70 R4G2Z0 A0A069DPA1 T1J0V0 A0A1B6M5T1 A0A0L7RII3 A0A2P6KL38 A0A0B6ZEJ4 A0A2A3ECX9 V9IC93 A0A088ARK8 B4MZZ5 A0A154P616 A0A0L8GUA9 T1FZF6 A0A1B6CL54 A0A1Y1LNE1 A0A0M9A7S8 A0A0P4XG69 A0A0N8BF97 A0A1Y1LNP4 A0A164Z295 A0A0P5FH00 A0A3B4CRS2 B4GSB4 Q29KG1 A0A2H8TLA3 A0A2S2NC48 A0A3S1BQ87 C4WXE2 A0A0K8UGC6 B4M9I3 A0A3B5QXW0 A0A3B5KTG5 A0A1S3DR92 A0A1L8DGG3 A0A1L8DG98 A0A2P2IA21 A0A2R7X890 A0A1W4UTH2 A0A2S2QJH9 A0A3B0JND8 T1HHZ1 B4Q8M7 Q8IPZ7 A0A1S3D5N1 B3NAM4 A0A3B5K1M6 V4BNL3 Q4V620 A0A3B1JU85 B0XE29 A0A3P8YUD7
A0A194RRS0 A0A067RL84 R4UW31 A0A0C9S9Z9 A0A194ANC0 K7INW7 A0A232FLP9 U4UG11 N6SYY1 A0A210PXR8 A0A131YR78 A0A224YST4 A0A131XQZ5 A0A195EU77 F4W485 A0A158NFL2 A0A195E4R7 A0A195B3H4 A0A1B6LDJ9 A0A146LJY8 A0A0P4VQ73 A0A146LUS3 A0A0A9Y3E5 A0A151X1M5 A0A195CKE6 A0A026VVZ2 A0A087T6F7 A0A1E1WW84 A0A2R5LES3 A0A147BF49 A0A0T6B2R1 A0A1B6JYY9 A0A1S3I4G4 A0A224XNN6 D1ZZY7 A0A0P4VL70 R4G2Z0 A0A069DPA1 T1J0V0 A0A1B6M5T1 A0A0L7RII3 A0A2P6KL38 A0A0B6ZEJ4 A0A2A3ECX9 V9IC93 A0A088ARK8 B4MZZ5 A0A154P616 A0A0L8GUA9 T1FZF6 A0A1B6CL54 A0A1Y1LNE1 A0A0M9A7S8 A0A0P4XG69 A0A0N8BF97 A0A1Y1LNP4 A0A164Z295 A0A0P5FH00 A0A3B4CRS2 B4GSB4 Q29KG1 A0A2H8TLA3 A0A2S2NC48 A0A3S1BQ87 C4WXE2 A0A0K8UGC6 B4M9I3 A0A3B5QXW0 A0A3B5KTG5 A0A1S3DR92 A0A1L8DGG3 A0A1L8DG98 A0A2P2IA21 A0A2R7X890 A0A1W4UTH2 A0A2S2QJH9 A0A3B0JND8 T1HHZ1 B4Q8M7 Q8IPZ7 A0A1S3D5N1 B3NAM4 A0A3B5K1M6 V4BNL3 Q4V620 A0A3B1JU85 B0XE29 A0A3P8YUD7
PDB
3CW1
E-value=4.15747e-09,
Score=139
Ontologies
PANTHER
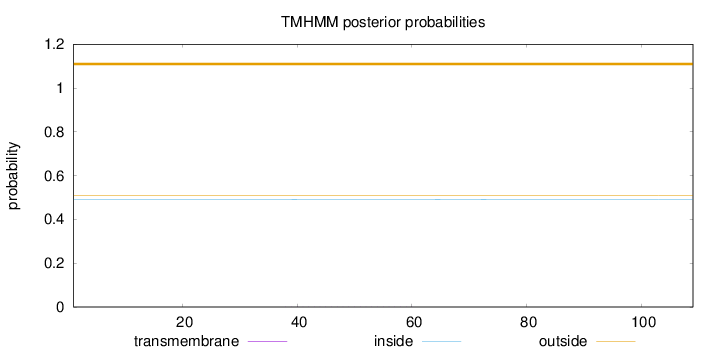
Topology
Length:
109
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01125
Exp number, first 60 AAs:
0.00944
Total prob of N-in:
0.49198
outside
1 - 109
Population Genetic Test Statistics
Pi
171.829115
Theta
182.224901
Tajima's D
-0.885575
CLR
0.336348
CSRT
0.15949202539873
Interpretation
Uncertain
