Gene
KWMTBOMO10919
Annotation
PREDICTED:_uncharacterized_protein_LOC105392935_[Plutella_xylostella]
Location in the cell
Nuclear Reliability : 3.558
Sequence
CDS
ATGGCAAGACGACGATCAAAAAGCCAGTCCTTCGATGACGACGACAGTATTTTCGATCGACCAACGCGTGGTGGAATGTTTCACTTAACGGAGAATATCATCAGGATGTTTGACGGCAAAGACAAAACTTATGCTGTAACAAAGTGGATACAAGATATAGAGGAAAATGGTGACATATTCGAGTGGACACCAAAGCAACGGTTATTAGTGGCTCGACGGTCATTAACCGGCACGGCAGCTTTGTGGTTAAATTCTGAAAAAACGTTTCCATGCTGGGATACTTTAAAGATAGCCATCAGTAAGGAGTTCCCTGACACCATAGACGCCAAGACTATTCACGAAATGATGAGTGCTAAAAAGAAAAAGTCAAATGAATCATGCATTGACTACATGTTTGCCATGAAAGAGCTTGGACGCCGAGGCAAAATGGCGGACTACGTGGCTATAAAATATGTTGTAGATGGTATACAAGATCAAGAAATAAATAAAATAATGCTATATGGAGTCACAACGTACGCTGAATTAAAGGAGAAATTAAAAATATACGAGATTTTGAAAGAAAAAATGAAGAAAGAACCTATTCACGGAAAAGAGCAGAAAATTACTAATACAAGAGAATGGTCATCAAAAGTTCGATGTTATAGCTGTGGAGAAAAGAGTCATAAAAGTGAAGATTGTCCGAAAAAGATGCAAGGAAAAAAGTGTTTCAACTGCAACGTGTTCGGCCATATTTCTTCACAATGTCCGGCAAAGGCGGAAGCGTCATCGGCTACGGCTAGCAGCGCGAGAACGGAGCAGCAACCGAAACGCACATATATCGCTACGACGCAGGAGGGCGCGAACACAATTTTCGAAAATCAATCTAACGAAGTTCACAACGCTATGAATGCTGTGATTGGCTCTGCGAACAGTACGGTGCGCGACAAATGTCATAACAGTGAGGTTGACATTTTAAAAGTAAATAAAGAATGTAAAGTGTTGAAAAAAACTTTGAAATTAGTAAAATTGTGTAATAATAACGAAATTCATGCATTAATTGATACTGGTAGTGAAGTTAATTTAGTAACCATGGACTATTATGAGCAAATAGGCTCTCCGACGTGTTTTAAAAATGTAAACACAGTTCTTACCGGTTTAGGCTCATCAAAGGTAATATCAAGTGAACAATTTAAAACAAACATTGAAATCGACGGCTACACGTATCGTGATATAATATTTTATGTGATTCCTAGAGACAGTATACCTATTAAAGTGATATTGGGACATGAATTTTTAAGAAATGTTATTACAGTTATGAAAGATGGTTTGGTATGGATGAAACCAAATGATTGGTTGTTTAAGGTAAATTGTTTTCCATGTTCTATTGACATTGTAGGTCATGTTATGAATGAAAATGTGAGGAATGAAGTGACTAGGTTAGTAGATGAATATCGGCCATGTCAAACTAAAGAGGCTCCCATTGAGTTGAAAATCACACTTAAAGACGACATCCCAGTGGCTCAACGACCACGACGCATCTCACCAAAAGAACAGGATGAGGAGGATTACTCTGCAAGGGACCACAACATTGTTTGGTTGTACCACAGAAGATGGAAATTGAAATTATTAAGAGGGCGCATGATAATGGTCATTTTTCAAAGAAGAAAACAATGGAATTGA
Protein
MARRRSKSQSFDDDDSIFDRPTRGGMFHLTENIIRMFDGKDKTYAVTKWIQDIEENGDIFEWTPKQRLLVARRSLTGTAALWLNSEKTFPCWDTLKIAISKEFPDTIDAKTIHEMMSAKKKKSNESCIDYMFAMKELGRRGKMADYVAIKYVVDGIQDQEINKIMLYGVTTYAELKEKLKIYEILKEKMKKEPIHGKEQKITNTREWSSKVRCYSCGEKSHKSEDCPKKMQGKKCFNCNVFGHISSQCPAKAEASSATASSARTEQQPKRTYIATTQEGANTIFENQSNEVHNAMNAVIGSANSTVRDKCHNSEVDILKVNKECKVLKKTLKLVKLCNNNEIHALIDTGSEVNLVTMDYYEQIGSPTCFKNVNTVLTGLGSSKVISSEQFKTNIEIDGYTYRDIIFYVIPRDSIPIKVILGHEFLRNVITVMKDGLVWMKPNDWLFKVNCFPCSIDIVGHVMNENVRNEVTRLVDEYRPCQTKEAPIELKITLKDDIPVAQRPRRISPKEQDEEDYSARDHNIVWLYHRRWKLKLLRGRMIMVIFQRRKQWN
Summary
Uniprot
EMBL
NWSH01001586
PCG70783.1
ODYU01001505
SOQ37806.1
GBBI01004550
JAC14162.1
+ More
GBBI01004551 JAC14161.1 LBMM01010976 KMQ87102.1 GBBI01004477 JAC14235.1 GDHF01023027 JAI29287.1 LBMM01004258 KMQ92652.1 ABLF02005846 NNAY01005459 OXU16662.1 GBGD01000341 JAC88548.1 GFTR01008753 JAW07673.1 GGFK01005990 MBW39311.1 GDHF01031400 JAI20914.1 GAMC01015907 JAB90648.1 GBXI01004632 JAD09660.1 LBMM01005919 KMQ91095.1
GBBI01004551 JAC14161.1 LBMM01010976 KMQ87102.1 GBBI01004477 JAC14235.1 GDHF01023027 JAI29287.1 LBMM01004258 KMQ92652.1 ABLF02005846 NNAY01005459 OXU16662.1 GBGD01000341 JAC88548.1 GFTR01008753 JAW07673.1 GGFK01005990 MBW39311.1 GDHF01031400 JAI20914.1 GAMC01015907 JAB90648.1 GBXI01004632 JAD09660.1 LBMM01005919 KMQ91095.1
Proteomes
Pfam
Interpro
IPR021109
Peptidase_aspartic_dom_sf
+ More
IPR036875 Znf_CCHC_sf
IPR001878 Znf_CCHC
IPR001969 Aspartic_peptidase_AS
IPR001995 Peptidase_A2_cat
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
IPR041373 RT_RNaseH
IPR000477 RT_dom
IPR041588 Integrase_H2C2
IPR036397 RNaseH_sf
IPR036361 SAP_dom_sf
IPR041577 RT_RNaseH_2
IPR003034 SAP_dom
IPR005162 Retrotrans_gag_dom
IPR036875 Znf_CCHC_sf
IPR001878 Znf_CCHC
IPR001969 Aspartic_peptidase_AS
IPR001995 Peptidase_A2_cat
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
IPR041373 RT_RNaseH
IPR000477 RT_dom
IPR041588 Integrase_H2C2
IPR036397 RNaseH_sf
IPR036361 SAP_dom_sf
IPR041577 RT_RNaseH_2
IPR003034 SAP_dom
IPR005162 Retrotrans_gag_dom
Gene 3D
ProteinModelPortal
PDB
2CQF
E-value=0.00117484,
Score=101
Ontologies
GO
Topology
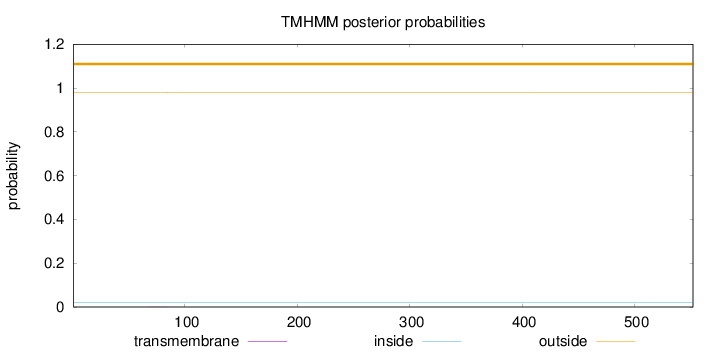
Length:
552
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0055
Exp number, first 60 AAs:
0.00013
Total prob of N-in:
0.01962
outside
1 - 552
Population Genetic Test Statistics
Pi
231.311252
Theta
150.823258
Tajima's D
-0.553221
CLR
129.528336
CSRT
0.230388480575971
Interpretation
Uncertain
