Gene
KWMTBOMO10917
Annotation
PREDICTED:_uncharacterized_protein_LOC105396011_[Plutella_xylostella]
Location in the cell
Nuclear Reliability : 3.616
Sequence
CDS
ATGGATGAACTTTTGAATGTTTTAGAAGATACCGCTGCGCTCTTAAACAAAACTCAAATTAATTTAAAGAAATGTCCAAAAGCTAGATTGACAAGAGGTTATGTAGAAGCAAGAATCAAATGTATCGAAGAATATTGGACTACATTTAAGCAAACGCATCTACAATTGGTAAAAATAACACCGAAAGAACAAAGACAAGAGTTACCATATTTTCAAAATGAAGATTACTTCGTGTTCGAAGATTTGTATCTCGTCTTGTTAGCCGATTTAAAAGATCTTTTGCAATCTTTCACACAAGCAGCCGCGCCGAGTACTTTAACGTCCTCACCAAATATTTCTAAGATTATTGCGGATACCGAAAACCTTGTTCGGTTACCGAGAATCGAATTACCTAAATTCACTGGAAACTATGAAGATTGGCCTACGTTTCAAGACCTATTTACCTCCTTAGTGGATAAGAACCCGGCTTTAAGTGACGTACAAAAGTTACATTATCTTAAGAGTACAGTCAGCTGCGAGGCAGAAACATTATTAAAACCTGTTCAAGTCACGGAAAACAATTATAAAAAAGCATGGAATATTTTGAAGTCAAGGTATGGAAATAAAAGATTAATAATTAACTCAGTTTTGAAGAAATTATTTAATCAGAAGAAAATTAGTTCTCAGTCTGCAAATCAATTAAAAAATTTATTAGACGTCACGACAGATTGTTTGAATAATTTAGAAAATTTAAATATAGCTACGAGCTCCTGGGATCCTATAATTATATTCCTTGTGATACAGAAGTTGGACCCGGAGTCTCACAAAGAATGGGAGCAGACAGCTTATTCAAAATCAGAGGACGAACTACCAACTTGGGAGAATTTAAAAGATTTCCTGCAGTCAAGATACCGTACCTTAGAACTTGTATTGTCCGTGTCAGGGACTTGTAGCAGCCGTGACAAACCTGTAAGAGAACGTTCACACTTAGTCACTGCCACAGCTACCACGTCACAGCCAAATGAAAGAACTTGCGTGCTCTGTAAAGAAAAGCATACATTATGTCATTGTAAGAATTTTACCACACTGCAACCTACAAAAAGAGCTGATTATGTCAAGACCAAAAATCTGTGTTTTAACTGTTTGGTGCCTGGGCATTCTGCTTACCAGTGTAAGTTACAAATGTCCTGTCGCCTATGTAGAAGACGACATCACACTCTTCTCCACCGCCATCAAGACTCGGCACAAACTGAGGAAAGGAATCAACCGAGTGCATCTCACCACGGGGTAGAAGAGAAAGAAGAGGTTATAAATAGCATGGTGGCGTCACATCACAGTACTAAACAAAGCACAGCACTACTAGCCACAGCGATGGTTTTAGCGAGGAACGAGCATAACCAAACCGTTGTTCTTCGTGCGCTAGTAGACCAAGGTTCGCAGGCTTCATTTTTAAGCGAGAAGGCGGCTCAAATGCTCAAACTTAACAGGCAACCAGCAAGAGGAAGTATCATCGGGGTAGGATCTACTAGGACTAACGTTGATCACATGGTTCAACTTAAGATTGGCTCCCGCTGGAATCCAAGCTTCGAAATCAACATACAAGCATATGTTATGAGTAAACAGTTGACCACGAAGATCCCCTCCAAAGCTGTAACTGTCACACATTGGCCACATCTAGAAGGACTCAACCTGGCAGATCCAGACTACTTCCAACCAGGTTCTATAGACTTACTTCTTGGTGTTAAAGAGTATGCACAGATTGTGCAACCAACACTTATTAAGGGCCCTCCAGGTTCTCCGTGTGCCCAAGAAACTAACCTGGGGTGGATCCTCTTCGGGGAAATCGATAATAACCGTGAAAATGAATCATTTCTGGTCCTTCATCATCAAGTCCATATAGATCATATGTTGAAATCCCTCTGGGAAATTGACAATGATAAAAAGAGAATATTAACAAAGGAAGAAAAGCTATGTGAAACAATATATGAAAAAACACATACGAGAAATAGTGAAGGAAGATATCTAGTAAGGCTGCCATTTAATACAAATAATCCAATTTCGACTGAAGGAAATACGAAAGAAATAGCTAGAAAAAGGTTACTTCAACTAGAAAGAAGATTTAAGAAAGAAACTAAACTGAAAGAAGATTATACAAAGGTCATGCAAGAATACATAAAAGCTAACTACATGGAAGAAATACCTAAGAATGAAATAAACAAGAAAAAATCAGTATACTTACCTCATCACGCCGTGGTACACGCAGACAGAGAAACTTCTAAGACACGCGTAGTATTTGATGCTTCAGCGAAGGGCTCTAACTGCGTTTCTCTAAACGATGAATTGCTTGTAGGTCCGCAACTGCAAGAGGACTTAAGGAACATACTTATGAGATCGAGATTACATCGTGTTTGCTATGCAAGTGATATACAAAAGATGTACCTGCAAATATTACTTTATGAAGAAGACGCCGATCGATATCAACGCCTTTTGTGGAGAGAGAATGATTCAGACCCGATTAAAGAATATCGTATGCTTCGTGTTACATTTGGTACAGCGGCGGCCCCTCACCTGGCTGTAAGAACGCTTCACCAGGTAGCTGACGATGAGGGTCGAAATCATCCAATGGCTGTTCGAATTATTAAAGAAGATTTTTACATGGACGATCTAATGTCGGGGGAGAGCAGTGCTCCTGAAACCATAAAGAGAGCACAAGAAGTAGATAATATTCTACAGAAAGGTGGATTTGTATTATCAAAATGGTCATCCAATTCTGCTGAATTTTTGAAATCTATCGAACCAAACAAAAGGACATCTCGAGCACAGTTGGATTTTAAATTGGATGGAAAAGTACAAGCTCTAGGTTTGATCTGGAACCTGGGCACGGATCAGTTTCAGTACAAGGTTAACTTACCAGCAAAGTCAAGCAGTATTACAAAGAGAGGCATACTATCTGATATTCAAAGACTTTTTGACCCACTGGGATGGATCGCCCCTAGCTTAATTTTGGCTAAGATACTCATACAAAGGCTATGGCTAGAACCGATTGGCTGGGATGACGAGATTAACCAATCGCTGTTACACGATTGGCTGACAATTAGAGAAGACTTTGAACATGTAAAAGAGATAGGTATTCCAAGATGGCTAAACACAGTAAGCACACAGATGGAAAACATCGAGGTTCATGGTTTCAGTGATGCTTCCATGAAAGCCTATGCCGCGGTGGCTTACATTCGTGTGAAGAGGGAAGATGGTTCAGTATTCACCAATCTATTAGCTGCCCGTACTAGAGTGGCACCACTCCGTACCGTGTCTCTACCACGCCTGGAGTTATGTGGGGCGCTGTTATTAGCAAGGTTACTTAAGCAAATAGAAAATGCAATGCGTATACCTACTTCTAACATCTATGCATGGACAGATTCATCAATTGTTCTTGCATGGCTATCTGGTGATCCTCATAGATGGAAGACATTCGTGGCGAACCGGGTCGTAGAAATTGTAGAAAATGTAAACACACACCGATGGTTCCATGTAGCTTCGGAAGAGAATCCTGCAGACATCGGGTCGCGGGGCATGCTGCTGGCGGACTTGAAACAGAAAGAGTTATGGTGGACAGGGCCCAAGTGGTTATGCAAAAAAGAAATAATATACAGTAAGCCAGGAATAACAGAAACAGAATTAGAAATGAAAACAATAAAATTAAATACTTACCACAATGCAGAAGAACTAAAACCTTTAACCGCTCAGTTTGAAGATTTTGATACGTTACCAGAATTACTAAAAACTATTATTTACTGTAAAAGATTTCTAAGATACAAAAGTAATTTAAGTACGAGCAAGGAAATAACTACACAAGAATTAGAAGACGCGCTTATCACATGCATTAAGTTAGTACAACAAGAATTCAAAGAAGAAATAGATCGGCTAAGAGATAAGAAACAAGTTAGAAGAAAAAGTTTCAACAAAAGGTCGGATTAA
Protein
MDELLNVLEDTAALLNKTQINLKKCPKARLTRGYVEARIKCIEEYWTTFKQTHLQLVKITPKEQRQELPYFQNEDYFVFEDLYLVLLADLKDLLQSFTQAAAPSTLTSSPNISKIIADTENLVRLPRIELPKFTGNYEDWPTFQDLFTSLVDKNPALSDVQKLHYLKSTVSCEAETLLKPVQVTENNYKKAWNILKSRYGNKRLIINSVLKKLFNQKKISSQSANQLKNLLDVTTDCLNNLENLNIATSSWDPIIIFLVIQKLDPESHKEWEQTAYSKSEDELPTWENLKDFLQSRYRTLELVLSVSGTCSSRDKPVRERSHLVTATATTSQPNERTCVLCKEKHTLCHCKNFTTLQPTKRADYVKTKNLCFNCLVPGHSAYQCKLQMSCRLCRRRHHTLLHRHQDSAQTEERNQPSASHHGVEEKEEVINSMVASHHSTKQSTALLATAMVLARNEHNQTVVLRALVDQGSQASFLSEKAAQMLKLNRQPARGSIIGVGSTRTNVDHMVQLKIGSRWNPSFEINIQAYVMSKQLTTKIPSKAVTVTHWPHLEGLNLADPDYFQPGSIDLLLGVKEYAQIVQPTLIKGPPGSPCAQETNLGWILFGEIDNNRENESFLVLHHQVHIDHMLKSLWEIDNDKKRILTKEEKLCETIYEKTHTRNSEGRYLVRLPFNTNNPISTEGNTKEIARKRLLQLERRFKKETKLKEDYTKVMQEYIKANYMEEIPKNEINKKKSVYLPHHAVVHADRETSKTRVVFDASAKGSNCVSLNDELLVGPQLQEDLRNILMRSRLHRVCYASDIQKMYLQILLYEEDADRYQRLLWRENDSDPIKEYRMLRVTFGTAAAPHLAVRTLHQVADDEGRNHPMAVRIIKEDFYMDDLMSGESSAPETIKRAQEVDNILQKGGFVLSKWSSNSAEFLKSIEPNKRTSRAQLDFKLDGKVQALGLIWNLGTDQFQYKVNLPAKSSSITKRGILSDIQRLFDPLGWIAPSLILAKILIQRLWLEPIGWDDEINQSLLHDWLTIREDFEHVKEIGIPRWLNTVSTQMENIEVHGFSDASMKAYAAVAYIRVKREDGSVFTNLLAARTRVAPLRTVSLPRLELCGALLLARLLKQIENAMRIPTSNIYAWTDSSIVLAWLSGDPHRWKTFVANRVVEIVENVNTHRWFHVASEENPADIGSRGMLLADLKQKELWWTGPKWLCKKEIIYSKPGITETELEMKTIKLNTYHNAEELKPLTAQFEDFDTLPELLKTIIYCKRFLRYKSNLSTSKEITTQELEDALITCIKLVQQEFKEEIDRLRDKKQVRRKSFNKRSD
Summary
Uniprot
A0A2H1WH28
A0A3S2L012
A0A0N1IHU1
A0A0N0PAG9
A0A1Y1LBZ3
A0A0J7N890
+ More
A0A226D417 A0A226E226 A0A0J7KCB9 A0A0J7KFC5 W8CCT3 A0A0J7N314 A0A226DMG9 A0A0J7N2B8 A0A0J7KBZ0 A0A0J7N465 A0A0J7K907 A0A1I8M1N7 A0A226EUF5 A0A0J7KB34 A0A0J7KCR4 A0A0J7KGI8 A0A0J7K9A7 A0A226E0X4 A0A226EM86 A0A226DKA3 A0A226EUK6 A0A0J7KBS8 A0A226D0T3 A0A0J7N189 Q8MNR4 A0A2A4K6T9 A0A0J7KCI2 J9LHB7 X1XTY9 A0A224XG39 A0A226DBN6 A0A023EZB2 A0A023EZC2 A0A226EPI5 A0A087T9T6 A0A226EAM0 A0A2S2PCB8 A0A2S2PNZ4 J9K774 J9LVB4 A0A0J7KGQ6 A0A2S2PTX9 J9LPA9 A0A2H1X238 J9JT64 J9LYM7 X1WY95 X1WRA3 A0A1W7R6S6 A0A1Y1N4T1 J9JVD6 J9KCB6 A0A2S2P9U8 J9JNP4 A0A2W1BJC9 X1WNM7 Q4JS97 A7XDH0 A0A2S2QPS5 A0A1W7R6I7 A0A194RAG7 A0A2S2PUJ2 J9KBH1 A0A2S2QB76 J9K4R2 A0A226D8N0 X1XU26 A0A023F0J1 A0A0J7KDN6 A0A1X7UAA5 X1WN86 X1XTX4 A0A1S3DJJ8 A0A023F0C4 A0A023EYR9 X1WVV7 W8ANH7 A0A2S2QQM0 A0A2S2NCU7 A0A1Y1L369 A0A1W7R6J0 A0A182H647 A0A2S2PLU2 A0A1S3DJC0 A0A034WA88 A0A1S3DJJ0 A0A1S3DJ34 A0A0J7K7U0 A0A355B931 A0A226DS81 W8C1U1 V5GU69
A0A226D417 A0A226E226 A0A0J7KCB9 A0A0J7KFC5 W8CCT3 A0A0J7N314 A0A226DMG9 A0A0J7N2B8 A0A0J7KBZ0 A0A0J7N465 A0A0J7K907 A0A1I8M1N7 A0A226EUF5 A0A0J7KB34 A0A0J7KCR4 A0A0J7KGI8 A0A0J7K9A7 A0A226E0X4 A0A226EM86 A0A226DKA3 A0A226EUK6 A0A0J7KBS8 A0A226D0T3 A0A0J7N189 Q8MNR4 A0A2A4K6T9 A0A0J7KCI2 J9LHB7 X1XTY9 A0A224XG39 A0A226DBN6 A0A023EZB2 A0A023EZC2 A0A226EPI5 A0A087T9T6 A0A226EAM0 A0A2S2PCB8 A0A2S2PNZ4 J9K774 J9LVB4 A0A0J7KGQ6 A0A2S2PTX9 J9LPA9 A0A2H1X238 J9JT64 J9LYM7 X1WY95 X1WRA3 A0A1W7R6S6 A0A1Y1N4T1 J9JVD6 J9KCB6 A0A2S2P9U8 J9JNP4 A0A2W1BJC9 X1WNM7 Q4JS97 A7XDH0 A0A2S2QPS5 A0A1W7R6I7 A0A194RAG7 A0A2S2PUJ2 J9KBH1 A0A2S2QB76 J9K4R2 A0A226D8N0 X1XU26 A0A023F0J1 A0A0J7KDN6 A0A1X7UAA5 X1WN86 X1XTX4 A0A1S3DJJ8 A0A023F0C4 A0A023EYR9 X1WVV7 W8ANH7 A0A2S2QQM0 A0A2S2NCU7 A0A1Y1L369 A0A1W7R6J0 A0A182H647 A0A2S2PLU2 A0A1S3DJC0 A0A034WA88 A0A1S3DJJ0 A0A1S3DJ34 A0A0J7K7U0 A0A355B931 A0A226DS81 W8C1U1 V5GU69
Pubmed
EMBL
ODYU01008608
SOQ52337.1
RSAL01000327
RVE42512.1
KQ460036
KPJ18393.1
+ More
KQ458473 KPJ05885.1 GEZM01060254 JAV71154.1 LBMM01008464 KMQ88855.1 LNIX01000036 OXA39969.1 LNIX01000007 OXA51795.1 LBMM01009645 KMQ87967.1 LBMM01008324 KMQ88972.1 GAMC01003146 JAC03410.1 LBMM01011051 KMQ87060.1 LNIX01000016 OXA46198.1 LBMM01011492 KMQ86800.1 LBMM01009964 KMQ87756.1 LBMM01010417 KMQ87475.1 LBMM01011516 KMQ86784.1 LNIX01000002 OXA60848.1 LBMM01010405 KMQ87487.1 LBMM01009551 KMQ88039.1 LBMM01007859 KMQ89359.1 LBMM01011283 KMQ86917.1 OXA51382.1 LNIX01000003 OXA58579.1 LNIX01000018 OXA45284.1 LNIX01000001 OXA61293.1 LBMM01009888 KMQ87813.1 LNIX01000044 OXA38660.1 LBMM01012077 KMQ86420.1 AJ487856 CAD32253.1 NWSH01000070 PCG79985.1 LBMM01009688 KMQ87939.1 ABLF02022040 ABLF02056888 ABLF02022912 ABLF02038819 ABLF02051942 ABLF02060282 GFTR01008874 JAW07552.1 LNIX01000026 OXA42254.1 GBBI01004140 JAC14572.1 GBBI01004139 JAC14573.1 OXA59522.1 KK114194 KFM61875.1 LNIX01000005 OXA54553.1 OXA54559.1 GGMR01014413 MBY27032.1 GGMR01018506 MBY31125.1 ABLF02023723 ABLF02004142 LBMM01007770 KMQ89434.1 GGMR01020322 MBY32941.1 ABLF02035780 ODYU01012875 SOQ59395.1 ABLF02023842 ABLF02028949 ABLF02058391 ABLF02016109 ABLF02016113 ABLF02030854 ABLF02067347 ABLF02066034 GEHC01000870 JAV46775.1 GEZM01012999 JAV92824.1 ABLF02008594 ABLF02042264 GGMR01013586 MBY26205.1 ABLF02005684 KZ150008 PZC75149.1 ABLF02042806 CR954257 CAJ14165.1 EF426679 ABP48078.1 GGMS01010524 MBY79727.1 GEHC01000869 JAV46776.1 KQ460500 KPJ14270.1 GGMR01020500 MBY33119.1 ABLF02018588 ABLF02018590 ABLF02042430 GGMS01005792 MBY74995.1 ABLF02007122 ABLF02033517 LNIX01000027 OXA41905.1 ABLF02001506 ABLF02008029 ABLF02035899 ABLF02037719 ABLF02046327 ABLF02064627 GBBI01004211 JAC14501.1 LBMM01009202 KMQ88311.1 ABLF02041590 ABLF02016099 GBBI01004408 JAC14304.1 GBBI01004410 JAC14302.1 ABLF02041505 GAMC01020552 JAB86003.1 GGMS01010815 MBY80018.1 GGMR01002340 MBY14959.1 GEZM01066272 JAV68024.1 GEHC01000877 JAV46768.1 JXUM01113346 KQ565681 KXJ70776.1 GGMR01017803 MBY30422.1 GAKP01007887 JAC51065.1 LBMM01012238 KMQ86334.1 DNYX01000039 HBK82233.1 LNIX01000012 OXA48073.1 GAMC01006864 JAB99691.1 GALX01003264 JAB65202.1
KQ458473 KPJ05885.1 GEZM01060254 JAV71154.1 LBMM01008464 KMQ88855.1 LNIX01000036 OXA39969.1 LNIX01000007 OXA51795.1 LBMM01009645 KMQ87967.1 LBMM01008324 KMQ88972.1 GAMC01003146 JAC03410.1 LBMM01011051 KMQ87060.1 LNIX01000016 OXA46198.1 LBMM01011492 KMQ86800.1 LBMM01009964 KMQ87756.1 LBMM01010417 KMQ87475.1 LBMM01011516 KMQ86784.1 LNIX01000002 OXA60848.1 LBMM01010405 KMQ87487.1 LBMM01009551 KMQ88039.1 LBMM01007859 KMQ89359.1 LBMM01011283 KMQ86917.1 OXA51382.1 LNIX01000003 OXA58579.1 LNIX01000018 OXA45284.1 LNIX01000001 OXA61293.1 LBMM01009888 KMQ87813.1 LNIX01000044 OXA38660.1 LBMM01012077 KMQ86420.1 AJ487856 CAD32253.1 NWSH01000070 PCG79985.1 LBMM01009688 KMQ87939.1 ABLF02022040 ABLF02056888 ABLF02022912 ABLF02038819 ABLF02051942 ABLF02060282 GFTR01008874 JAW07552.1 LNIX01000026 OXA42254.1 GBBI01004140 JAC14572.1 GBBI01004139 JAC14573.1 OXA59522.1 KK114194 KFM61875.1 LNIX01000005 OXA54553.1 OXA54559.1 GGMR01014413 MBY27032.1 GGMR01018506 MBY31125.1 ABLF02023723 ABLF02004142 LBMM01007770 KMQ89434.1 GGMR01020322 MBY32941.1 ABLF02035780 ODYU01012875 SOQ59395.1 ABLF02023842 ABLF02028949 ABLF02058391 ABLF02016109 ABLF02016113 ABLF02030854 ABLF02067347 ABLF02066034 GEHC01000870 JAV46775.1 GEZM01012999 JAV92824.1 ABLF02008594 ABLF02042264 GGMR01013586 MBY26205.1 ABLF02005684 KZ150008 PZC75149.1 ABLF02042806 CR954257 CAJ14165.1 EF426679 ABP48078.1 GGMS01010524 MBY79727.1 GEHC01000869 JAV46776.1 KQ460500 KPJ14270.1 GGMR01020500 MBY33119.1 ABLF02018588 ABLF02018590 ABLF02042430 GGMS01005792 MBY74995.1 ABLF02007122 ABLF02033517 LNIX01000027 OXA41905.1 ABLF02001506 ABLF02008029 ABLF02035899 ABLF02037719 ABLF02046327 ABLF02064627 GBBI01004211 JAC14501.1 LBMM01009202 KMQ88311.1 ABLF02041590 ABLF02016099 GBBI01004408 JAC14304.1 GBBI01004410 JAC14302.1 ABLF02041505 GAMC01020552 JAB86003.1 GGMS01010815 MBY80018.1 GGMR01002340 MBY14959.1 GEZM01066272 JAV68024.1 GEHC01000877 JAV46768.1 JXUM01113346 KQ565681 KXJ70776.1 GGMR01017803 MBY30422.1 GAKP01007887 JAC51065.1 LBMM01012238 KMQ86334.1 DNYX01000039 HBK82233.1 LNIX01000012 OXA48073.1 GAMC01006864 JAB99691.1 GALX01003264 JAB65202.1
Proteomes
PRIDE
Pfam
Interpro
IPR001584
Integrase_cat-core
+ More
IPR012337 RNaseH-like_sf
IPR008042 Retrotrans_Pao
IPR001878 Znf_CCHC
IPR008737 Peptidase_asp_put
IPR040676 DUF5641
IPR005312 DUF1759
IPR041588 Integrase_H2C2
IPR036397 RNaseH_sf
IPR000253 FHA_dom
IPR000742 EGF-like_dom
IPR011042 6-blade_b-propeller_TolB-like
IPR013032 EGF-like_CS
IPR021109 Peptidase_aspartic_dom_sf
IPR000033 LDLR_classB_rpt
IPR000477 RT_dom
IPR001995 Peptidase_A2_cat
IPR001969 Aspartic_peptidase_AS
IPR000331 Rap_GAP_dom
IPR035974 Rap/Ran-GAP_sf
IPR000425 MIP
IPR023271 Aquaporin-like
IPR022048 Envelope_fusion-like
IPR000727 T_SNARE_dom
IPR041373 RT_RNaseH
IPR041577 RT_RNaseH_2
IPR036875 Znf_CCHC_sf
IPR012337 RNaseH-like_sf
IPR008042 Retrotrans_Pao
IPR001878 Znf_CCHC
IPR008737 Peptidase_asp_put
IPR040676 DUF5641
IPR005312 DUF1759
IPR041588 Integrase_H2C2
IPR036397 RNaseH_sf
IPR000253 FHA_dom
IPR000742 EGF-like_dom
IPR011042 6-blade_b-propeller_TolB-like
IPR013032 EGF-like_CS
IPR021109 Peptidase_aspartic_dom_sf
IPR000033 LDLR_classB_rpt
IPR000477 RT_dom
IPR001995 Peptidase_A2_cat
IPR001969 Aspartic_peptidase_AS
IPR000331 Rap_GAP_dom
IPR035974 Rap/Ran-GAP_sf
IPR000425 MIP
IPR023271 Aquaporin-like
IPR022048 Envelope_fusion-like
IPR000727 T_SNARE_dom
IPR041373 RT_RNaseH
IPR041577 RT_RNaseH_2
IPR036875 Znf_CCHC_sf
SUPFAM
ProteinModelPortal
A0A2H1WH28
A0A3S2L012
A0A0N1IHU1
A0A0N0PAG9
A0A1Y1LBZ3
A0A0J7N890
+ More
A0A226D417 A0A226E226 A0A0J7KCB9 A0A0J7KFC5 W8CCT3 A0A0J7N314 A0A226DMG9 A0A0J7N2B8 A0A0J7KBZ0 A0A0J7N465 A0A0J7K907 A0A1I8M1N7 A0A226EUF5 A0A0J7KB34 A0A0J7KCR4 A0A0J7KGI8 A0A0J7K9A7 A0A226E0X4 A0A226EM86 A0A226DKA3 A0A226EUK6 A0A0J7KBS8 A0A226D0T3 A0A0J7N189 Q8MNR4 A0A2A4K6T9 A0A0J7KCI2 J9LHB7 X1XTY9 A0A224XG39 A0A226DBN6 A0A023EZB2 A0A023EZC2 A0A226EPI5 A0A087T9T6 A0A226EAM0 A0A2S2PCB8 A0A2S2PNZ4 J9K774 J9LVB4 A0A0J7KGQ6 A0A2S2PTX9 J9LPA9 A0A2H1X238 J9JT64 J9LYM7 X1WY95 X1WRA3 A0A1W7R6S6 A0A1Y1N4T1 J9JVD6 J9KCB6 A0A2S2P9U8 J9JNP4 A0A2W1BJC9 X1WNM7 Q4JS97 A7XDH0 A0A2S2QPS5 A0A1W7R6I7 A0A194RAG7 A0A2S2PUJ2 J9KBH1 A0A2S2QB76 J9K4R2 A0A226D8N0 X1XU26 A0A023F0J1 A0A0J7KDN6 A0A1X7UAA5 X1WN86 X1XTX4 A0A1S3DJJ8 A0A023F0C4 A0A023EYR9 X1WVV7 W8ANH7 A0A2S2QQM0 A0A2S2NCU7 A0A1Y1L369 A0A1W7R6J0 A0A182H647 A0A2S2PLU2 A0A1S3DJC0 A0A034WA88 A0A1S3DJJ0 A0A1S3DJ34 A0A0J7K7U0 A0A355B931 A0A226DS81 W8C1U1 V5GU69
A0A226D417 A0A226E226 A0A0J7KCB9 A0A0J7KFC5 W8CCT3 A0A0J7N314 A0A226DMG9 A0A0J7N2B8 A0A0J7KBZ0 A0A0J7N465 A0A0J7K907 A0A1I8M1N7 A0A226EUF5 A0A0J7KB34 A0A0J7KCR4 A0A0J7KGI8 A0A0J7K9A7 A0A226E0X4 A0A226EM86 A0A226DKA3 A0A226EUK6 A0A0J7KBS8 A0A226D0T3 A0A0J7N189 Q8MNR4 A0A2A4K6T9 A0A0J7KCI2 J9LHB7 X1XTY9 A0A224XG39 A0A226DBN6 A0A023EZB2 A0A023EZC2 A0A226EPI5 A0A087T9T6 A0A226EAM0 A0A2S2PCB8 A0A2S2PNZ4 J9K774 J9LVB4 A0A0J7KGQ6 A0A2S2PTX9 J9LPA9 A0A2H1X238 J9JT64 J9LYM7 X1WY95 X1WRA3 A0A1W7R6S6 A0A1Y1N4T1 J9JVD6 J9KCB6 A0A2S2P9U8 J9JNP4 A0A2W1BJC9 X1WNM7 Q4JS97 A7XDH0 A0A2S2QPS5 A0A1W7R6I7 A0A194RAG7 A0A2S2PUJ2 J9KBH1 A0A2S2QB76 J9K4R2 A0A226D8N0 X1XU26 A0A023F0J1 A0A0J7KDN6 A0A1X7UAA5 X1WN86 X1XTX4 A0A1S3DJJ8 A0A023F0C4 A0A023EYR9 X1WVV7 W8ANH7 A0A2S2QQM0 A0A2S2NCU7 A0A1Y1L369 A0A1W7R6J0 A0A182H647 A0A2S2PLU2 A0A1S3DJC0 A0A034WA88 A0A1S3DJJ0 A0A1S3DJ34 A0A0J7K7U0 A0A355B931 A0A226DS81 W8C1U1 V5GU69
Ontologies
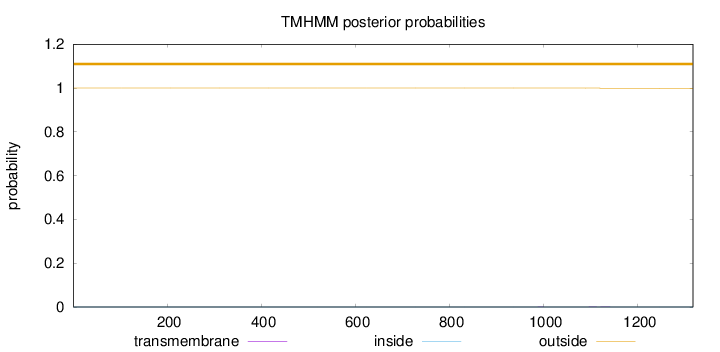
Topology
Length:
1317
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01071
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00001
outside
1 - 1317
Population Genetic Test Statistics
Pi
148.54495
Theta
163.274989
Tajima's D
-1.128125
CLR
0.138483
CSRT
0.121493925303735
Interpretation
Uncertain
