Gene
KWMTBOMO10916
Annotation
proteasome_alpha_3_subunit_[Bombyx_mori]
Full name
Proteasome subunit alpha type
+ More
Proteasome endopeptidase complex
Proteasome endopeptidase complex
Location in the cell
Cytoplasmic Reliability : 2.652
Sequence
CDS
ATGAGTTCTATCGGAACTGGCTATGACCTATCGGCTTCTCAATTTTCTCCTGATGGTCGCGTTTTCCAAGTGGAATATGCTGCAAAGGCTGTAGAAAATTCTGGAACAGTAATTGGCCTCCGAGGAAAAGACGGAGTGGTATTTGCTGTTGAGAAGTTAATCACATCCAAACTGTATGAACCTGGCGCAAACAAGAGGATATTCCATATTGATGAACATGTTGGCATGGCTGTCGCAGGGCTCATATCAGATGCAAGACAAATTGTCGAGACTGCTCGTTCAGAGGCATCAAACTACAGATCTCAGTATGGCAGTCCAGTACCACTGAAGTATTTAAATGAACGAGTTTCAATGTACATGCACGCATACACACTATACAGCGCTGTTCGGCCATATGGTTGCTCAGTCGTTATGGGCACATGGACAGACTATGAAGGCCCTCAGATGTATATGCTAGATCCCAGTGGTGTATCTTTTTCATATTTTGGTTGCGCAGTCGGAAAAGCCAAACAGGCAGCAAAAACAGAGATAGAAAAGCTAAAACTTGGAGACCTCACAGTGAAGGAGCTTGTGAGAGAAGCCGCTAGAATTATCTATCTTGTACATGATGAACTAAAAGATAAGCAGTTTGAGCTGGAGTTGTCTTGGGTTTCGAAAGACACGAAGGGTCGCCATGAGCTGGTTCCGCGTGAGCTCGCCACCGAAGCAGAGAACCAAGCTAAACAGGCTCTTGCTGATATTGAAGATTCAGATGAAGGAGAGATGTGA
Protein
MSSIGTGYDLSASQFSPDGRVFQVEYAAKAVENSGTVIGLRGKDGVVFAVEKLITSKLYEPGANKRIFHIDEHVGMAVAGLISDARQIVETARSEASNYRSQYGSPVPLKYLNERVSMYMHAYTLYSAVRPYGCSVVMGTWTDYEGPQMYMLDPSGVSFSYFGCAVGKAKQAAKTEIEKLKLGDLTVKELVREAARIIYLVHDELKDKQFELELSWVSKDTKGRHELVPRELATEAENQAKQALADIEDSDEGEM
Summary
Description
The proteasome is a multicatalytic proteinase complex which is characterized by its ability to cleave peptides with Arg, Phe, Tyr, Leu, and Glu adjacent to the leaving group at neutral or slightly basic pH.
Catalytic Activity
Cleavage of peptide bonds with very broad specificity.
Subunit
The 20S proteasome core is composed of 28 subunits that are arranged in four stacked rings, resulting in a barrel-shaped structure. The two end rings are each formed by seven alpha subunits, and the two central rings are each formed by seven beta subunits.
The 26S proteasome consists of a 20S proteasome core and two 19S regulatory subunits.
The 26S proteasome consists of a 20S proteasome core and two 19S regulatory subunits.
Similarity
Belongs to the peptidase T1A family.
Belongs to the G-protein coupled receptor 1 family.
Belongs to the UDP-glycosyltransferase family.
Belongs to the G-protein coupled receptor 1 family.
Belongs to the UDP-glycosyltransferase family.
Uniprot
Q1HQ44
A0A2A4J8C2
A0A2W1BDQ7
A0A194PN82
A0A212F8U7
A0A1E1WGR6
+ More
B2DBN8 S4PH61 A0A3S2L319 A0A0C9R691 A0A0N0BDS2 A0A2J7PWA0 A0A026W2X3 A0A158NGJ8 A0A151X1S9 E2BR97 A0A0T6B429 A0A0J7K1E1 E2ATL4 A0A1B6D194 A0A1W4WUU2 A0A023F9V8 A0A195DQJ6 A0A088AP06 E9IDY9 A0A195BXW8 A0A195EU44 A0A1Y1LYZ7 A0A1B1ZGH4 A0A212EQU0 A0A0P4VKU2 R4FNC4 A0A224XVW1 A0A0V0G5Y8 A0A1B6GTG8 A0A0A9YLA3 E9FYX7 A0A1B6MGG5 A0A2A3EQM0 D1ZZB6 A0A0P5IS80 A0A2P6K993 A0A0P5G3L5 A0A0P6CJV5 A0A0P5MPD7 A0A0P6CN59 A0A131Y0L0 B7PNN1 A0A0P5MR06 A0A0P5JUZ7 A0A0P5P768 A0A0P6G188 G9C5F2 T1JNT1 A0A0P6GJZ9 A0A023GGX0 A0A087SV27 G3MMG5 A0A0N8C7X5 A0A2R5LGB9 A0A0K8RKM3 A0A224YG46 A0A131YRB9 A0A131XMU9 A0A0P5N1M6 L7M7U4 A0A1Z5LBV0 A0A023FUV1 A0A3S4QKJ2 A0A226EQY9 A0A1E1X9Y7 A0A0L7QPU0 A0A293LHL9 A0A3R7SYE8 C3Y0B8 I1TLI5 C1BF68 B5XGI9 A0A0P4XYR5 A0A1W5AW99 A0A3B4DGN1 A0A2B4SGP9 W5MSR8 W5JYN0 A0A315W1F9 C1BLU4 I3JYD3 H2M390 A0A3P9LLQ8 J3JYN3 A0A2I0UFE9 E3TC48 A0A0P4VNE7 A0A194AM89 A0A3Q2CLV2 A0A3B3DWF1 A0A3P8VWA8 A0A060WGZ9 B5X5D8 A0A3S2NRP4
B2DBN8 S4PH61 A0A3S2L319 A0A0C9R691 A0A0N0BDS2 A0A2J7PWA0 A0A026W2X3 A0A158NGJ8 A0A151X1S9 E2BR97 A0A0T6B429 A0A0J7K1E1 E2ATL4 A0A1B6D194 A0A1W4WUU2 A0A023F9V8 A0A195DQJ6 A0A088AP06 E9IDY9 A0A195BXW8 A0A195EU44 A0A1Y1LYZ7 A0A1B1ZGH4 A0A212EQU0 A0A0P4VKU2 R4FNC4 A0A224XVW1 A0A0V0G5Y8 A0A1B6GTG8 A0A0A9YLA3 E9FYX7 A0A1B6MGG5 A0A2A3EQM0 D1ZZB6 A0A0P5IS80 A0A2P6K993 A0A0P5G3L5 A0A0P6CJV5 A0A0P5MPD7 A0A0P6CN59 A0A131Y0L0 B7PNN1 A0A0P5MR06 A0A0P5JUZ7 A0A0P5P768 A0A0P6G188 G9C5F2 T1JNT1 A0A0P6GJZ9 A0A023GGX0 A0A087SV27 G3MMG5 A0A0N8C7X5 A0A2R5LGB9 A0A0K8RKM3 A0A224YG46 A0A131YRB9 A0A131XMU9 A0A0P5N1M6 L7M7U4 A0A1Z5LBV0 A0A023FUV1 A0A3S4QKJ2 A0A226EQY9 A0A1E1X9Y7 A0A0L7QPU0 A0A293LHL9 A0A3R7SYE8 C3Y0B8 I1TLI5 C1BF68 B5XGI9 A0A0P4XYR5 A0A1W5AW99 A0A3B4DGN1 A0A2B4SGP9 W5MSR8 W5JYN0 A0A315W1F9 C1BLU4 I3JYD3 H2M390 A0A3P9LLQ8 J3JYN3 A0A2I0UFE9 E3TC48 A0A0P4VNE7 A0A194AM89 A0A3Q2CLV2 A0A3B3DWF1 A0A3P8VWA8 A0A060WGZ9 B5X5D8 A0A3S2NRP4
EC Number
3.4.25.1
Pubmed
28756777
26354079
22118469
18712529
23622113
24508170
+ More
30249741 21347285 20798317 25474469 21282665 28004739 27464714 27129103 25401762 26823975 21292972 18362917 19820115 21835214 22216098 28797301 26830274 28049606 25576852 28528879 28503490 18563158 22361323 24755649 20433749 25329095 29703783 25186727 17554307 22516182 20634964 29451363 24487278
30249741 21347285 20798317 25474469 21282665 28004739 27464714 27129103 25401762 26823975 21292972 18362917 19820115 21835214 22216098 28797301 26830274 28049606 25576852 28528879 28503490 18563158 22361323 24755649 20433749 25329095 29703783 25186727 17554307 22516182 20634964 29451363 24487278
EMBL
DQ443208
ABF51297.1
NWSH01002643
PCG67774.1
KZ150315
PZC71447.1
+ More
KQ459604 KPI92575.1 AGBW02009667 OWR50164.1 GDQN01004861 JAT86193.1 AB264723 BAG30797.1 GAIX01003382 JAA89178.1 RSAL01000198 RVE44527.1 GBYB01002261 JAG72028.1 KQ435859 KOX70598.1 NEVH01020936 PNF20606.1 KK107519 QOIP01000009 EZA49384.1 RLU18567.1 ADTU01001608 KQ982585 KYQ54363.1 GL449936 EFN81786.1 LJIG01015966 KRT82017.1 LBMM01016914 KMQ84268.1 GL442629 EFN63210.1 GEDC01017854 JAS19444.1 GBBI01000928 JAC17784.1 KQ980612 KYN15097.1 GL762556 EFZ21167.1 KQ976398 KYM92788.1 KQ981979 KYN31399.1 GEZM01045074 JAV77938.1 KU756280 ANX99823.1 AGBW02013211 OWR43863.1 GDKW01000877 JAI55718.1 ACPB03021727 GAHY01000623 JAA76887.1 GFTR01004183 JAW12243.1 GECL01003415 JAP02709.1 GECZ01004041 JAS65728.1 GBHO01011208 GBRD01013401 GDHC01014255 JAG32396.1 JAG52425.1 JAQ04374.1 GL732527 EFX87695.1 GEBQ01004964 JAT35013.1 KZ288193 PBC34007.1 KQ971338 EFA02345.1 GDIQ01210422 GDIQ01123550 LRGB01001151 JAK41303.1 KZS13437.1 MWRG01019710 PRD22879.1 GDIQ01246748 JAK04977.1 GDIQ01090163 JAN04574.1 GDIQ01155389 JAK96336.1 GDIQ01090162 JAN04575.1 GEFM01003791 JAP72005.1 ABJB010267734 DS753779 EEC08203.1 GDIQ01152589 JAK99136.1 GDIQ01194726 JAK56999.1 GDIQ01137001 JAL14725.1 GDIQ01040034 JAN54703.1 HQ851393 AEV89771.1 AFFK01014262 GDIQ01042682 JAN52055.1 GBBM01002201 JAC33217.1 KK112090 KFM56716.1 JO843066 AEO34683.1 GDIQ01106059 JAL45667.1 GGLE01004339 MBY08465.1 GADI01002173 JAA71635.1 GFPF01005500 MAA16646.1 GEDV01006753 JAP81804.1 GEFH01001700 JAP66881.1 GDIQ01152590 JAK99135.1 GACK01005806 JAA59228.1 GFJQ02002098 JAW04872.1 GBBL01001714 JAC25606.1 NCKU01005401 RWS04622.1 LNIX01000002 OXA60042.1 GFAC01003120 JAT96068.1 KQ414804 KOC60652.1 GFWV01008258 MAA32987.1 QCYY01001002 ROT81221.1 GG666478 EEN66191.1 JF834155 AFE88208.1 BT073247 FR904415 ACO07671.1 CDQ62692.1 BT050158 BT058648 BT060362 ACI69959.1 ACN10361.1 ACN12722.1 GDIP01235000 JAI88401.1 LSMT01000098 PFX27672.1 AHAT01003872 NHOQ01000541 PWA29759.1 BT075573 ACO09997.1 AERX01040125 AERX01040126 BT128362 AEE63320.1 KZ505805 PKU44768.1 GU587927 ADO27884.1 GDRN01112096 JAI56733.1 GELH01000598 GELH01000597 JAS03675.1 FR904536 CDQ66256.1 BT046257 ACI66058.1 CM012458 RVE57648.1
KQ459604 KPI92575.1 AGBW02009667 OWR50164.1 GDQN01004861 JAT86193.1 AB264723 BAG30797.1 GAIX01003382 JAA89178.1 RSAL01000198 RVE44527.1 GBYB01002261 JAG72028.1 KQ435859 KOX70598.1 NEVH01020936 PNF20606.1 KK107519 QOIP01000009 EZA49384.1 RLU18567.1 ADTU01001608 KQ982585 KYQ54363.1 GL449936 EFN81786.1 LJIG01015966 KRT82017.1 LBMM01016914 KMQ84268.1 GL442629 EFN63210.1 GEDC01017854 JAS19444.1 GBBI01000928 JAC17784.1 KQ980612 KYN15097.1 GL762556 EFZ21167.1 KQ976398 KYM92788.1 KQ981979 KYN31399.1 GEZM01045074 JAV77938.1 KU756280 ANX99823.1 AGBW02013211 OWR43863.1 GDKW01000877 JAI55718.1 ACPB03021727 GAHY01000623 JAA76887.1 GFTR01004183 JAW12243.1 GECL01003415 JAP02709.1 GECZ01004041 JAS65728.1 GBHO01011208 GBRD01013401 GDHC01014255 JAG32396.1 JAG52425.1 JAQ04374.1 GL732527 EFX87695.1 GEBQ01004964 JAT35013.1 KZ288193 PBC34007.1 KQ971338 EFA02345.1 GDIQ01210422 GDIQ01123550 LRGB01001151 JAK41303.1 KZS13437.1 MWRG01019710 PRD22879.1 GDIQ01246748 JAK04977.1 GDIQ01090163 JAN04574.1 GDIQ01155389 JAK96336.1 GDIQ01090162 JAN04575.1 GEFM01003791 JAP72005.1 ABJB010267734 DS753779 EEC08203.1 GDIQ01152589 JAK99136.1 GDIQ01194726 JAK56999.1 GDIQ01137001 JAL14725.1 GDIQ01040034 JAN54703.1 HQ851393 AEV89771.1 AFFK01014262 GDIQ01042682 JAN52055.1 GBBM01002201 JAC33217.1 KK112090 KFM56716.1 JO843066 AEO34683.1 GDIQ01106059 JAL45667.1 GGLE01004339 MBY08465.1 GADI01002173 JAA71635.1 GFPF01005500 MAA16646.1 GEDV01006753 JAP81804.1 GEFH01001700 JAP66881.1 GDIQ01152590 JAK99135.1 GACK01005806 JAA59228.1 GFJQ02002098 JAW04872.1 GBBL01001714 JAC25606.1 NCKU01005401 RWS04622.1 LNIX01000002 OXA60042.1 GFAC01003120 JAT96068.1 KQ414804 KOC60652.1 GFWV01008258 MAA32987.1 QCYY01001002 ROT81221.1 GG666478 EEN66191.1 JF834155 AFE88208.1 BT073247 FR904415 ACO07671.1 CDQ62692.1 BT050158 BT058648 BT060362 ACI69959.1 ACN10361.1 ACN12722.1 GDIP01235000 JAI88401.1 LSMT01000098 PFX27672.1 AHAT01003872 NHOQ01000541 PWA29759.1 BT075573 ACO09997.1 AERX01040125 AERX01040126 BT128362 AEE63320.1 KZ505805 PKU44768.1 GU587927 ADO27884.1 GDRN01112096 JAI56733.1 GELH01000598 GELH01000597 JAS03675.1 FR904536 CDQ66256.1 BT046257 ACI66058.1 CM012458 RVE57648.1
Proteomes
UP000218220
UP000053268
UP000007151
UP000283053
UP000053105
UP000235965
+ More
UP000053097 UP000279307 UP000005205 UP000075809 UP000008237 UP000036403 UP000000311 UP000192223 UP000078492 UP000005203 UP000078540 UP000078541 UP000015103 UP000000305 UP000242457 UP000007266 UP000076858 UP000001555 UP000054359 UP000285301 UP000198287 UP000053825 UP000283509 UP000001554 UP000193380 UP000087266 UP000192224 UP000261440 UP000225706 UP000018468 UP000018467 UP000005207 UP000001038 UP000265180 UP000265200 UP000265020 UP000261560 UP000265120
UP000053097 UP000279307 UP000005205 UP000075809 UP000008237 UP000036403 UP000000311 UP000192223 UP000078492 UP000005203 UP000078540 UP000078541 UP000015103 UP000000305 UP000242457 UP000007266 UP000076858 UP000001555 UP000054359 UP000285301 UP000198287 UP000053825 UP000283509 UP000001554 UP000193380 UP000087266 UP000192224 UP000261440 UP000225706 UP000018468 UP000018467 UP000005207 UP000001038 UP000265180 UP000265200 UP000265020 UP000261560 UP000265120
PRIDE
Interpro
SUPFAM
SSF56235
SSF56235
Gene 3D
ProteinModelPortal
Q1HQ44
A0A2A4J8C2
A0A2W1BDQ7
A0A194PN82
A0A212F8U7
A0A1E1WGR6
+ More
B2DBN8 S4PH61 A0A3S2L319 A0A0C9R691 A0A0N0BDS2 A0A2J7PWA0 A0A026W2X3 A0A158NGJ8 A0A151X1S9 E2BR97 A0A0T6B429 A0A0J7K1E1 E2ATL4 A0A1B6D194 A0A1W4WUU2 A0A023F9V8 A0A195DQJ6 A0A088AP06 E9IDY9 A0A195BXW8 A0A195EU44 A0A1Y1LYZ7 A0A1B1ZGH4 A0A212EQU0 A0A0P4VKU2 R4FNC4 A0A224XVW1 A0A0V0G5Y8 A0A1B6GTG8 A0A0A9YLA3 E9FYX7 A0A1B6MGG5 A0A2A3EQM0 D1ZZB6 A0A0P5IS80 A0A2P6K993 A0A0P5G3L5 A0A0P6CJV5 A0A0P5MPD7 A0A0P6CN59 A0A131Y0L0 B7PNN1 A0A0P5MR06 A0A0P5JUZ7 A0A0P5P768 A0A0P6G188 G9C5F2 T1JNT1 A0A0P6GJZ9 A0A023GGX0 A0A087SV27 G3MMG5 A0A0N8C7X5 A0A2R5LGB9 A0A0K8RKM3 A0A224YG46 A0A131YRB9 A0A131XMU9 A0A0P5N1M6 L7M7U4 A0A1Z5LBV0 A0A023FUV1 A0A3S4QKJ2 A0A226EQY9 A0A1E1X9Y7 A0A0L7QPU0 A0A293LHL9 A0A3R7SYE8 C3Y0B8 I1TLI5 C1BF68 B5XGI9 A0A0P4XYR5 A0A1W5AW99 A0A3B4DGN1 A0A2B4SGP9 W5MSR8 W5JYN0 A0A315W1F9 C1BLU4 I3JYD3 H2M390 A0A3P9LLQ8 J3JYN3 A0A2I0UFE9 E3TC48 A0A0P4VNE7 A0A194AM89 A0A3Q2CLV2 A0A3B3DWF1 A0A3P8VWA8 A0A060WGZ9 B5X5D8 A0A3S2NRP4
B2DBN8 S4PH61 A0A3S2L319 A0A0C9R691 A0A0N0BDS2 A0A2J7PWA0 A0A026W2X3 A0A158NGJ8 A0A151X1S9 E2BR97 A0A0T6B429 A0A0J7K1E1 E2ATL4 A0A1B6D194 A0A1W4WUU2 A0A023F9V8 A0A195DQJ6 A0A088AP06 E9IDY9 A0A195BXW8 A0A195EU44 A0A1Y1LYZ7 A0A1B1ZGH4 A0A212EQU0 A0A0P4VKU2 R4FNC4 A0A224XVW1 A0A0V0G5Y8 A0A1B6GTG8 A0A0A9YLA3 E9FYX7 A0A1B6MGG5 A0A2A3EQM0 D1ZZB6 A0A0P5IS80 A0A2P6K993 A0A0P5G3L5 A0A0P6CJV5 A0A0P5MPD7 A0A0P6CN59 A0A131Y0L0 B7PNN1 A0A0P5MR06 A0A0P5JUZ7 A0A0P5P768 A0A0P6G188 G9C5F2 T1JNT1 A0A0P6GJZ9 A0A023GGX0 A0A087SV27 G3MMG5 A0A0N8C7X5 A0A2R5LGB9 A0A0K8RKM3 A0A224YG46 A0A131YRB9 A0A131XMU9 A0A0P5N1M6 L7M7U4 A0A1Z5LBV0 A0A023FUV1 A0A3S4QKJ2 A0A226EQY9 A0A1E1X9Y7 A0A0L7QPU0 A0A293LHL9 A0A3R7SYE8 C3Y0B8 I1TLI5 C1BF68 B5XGI9 A0A0P4XYR5 A0A1W5AW99 A0A3B4DGN1 A0A2B4SGP9 W5MSR8 W5JYN0 A0A315W1F9 C1BLU4 I3JYD3 H2M390 A0A3P9LLQ8 J3JYN3 A0A2I0UFE9 E3TC48 A0A0P4VNE7 A0A194AM89 A0A3Q2CLV2 A0A3B3DWF1 A0A3P8VWA8 A0A060WGZ9 B5X5D8 A0A3S2NRP4
PDB
3UNH
E-value=2.687e-97,
Score=905
Ontologies
KEGG
GO
PANTHER
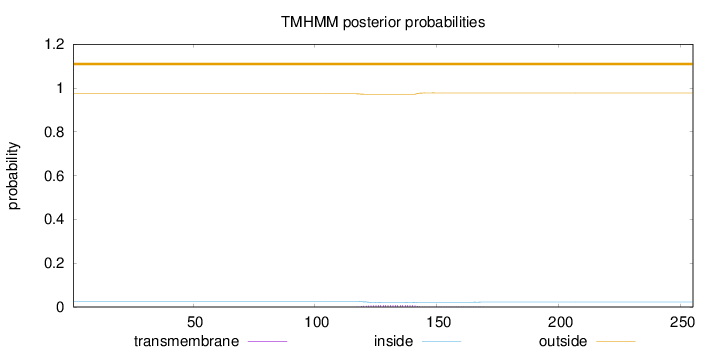
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleus
Length:
255
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.20305
Exp number, first 60 AAs:
7e-05
Total prob of N-in:
0.02642
outside
1 - 255
Population Genetic Test Statistics
Pi
242.341785
Theta
152.36064
Tajima's D
2.633676
CLR
0.885815
CSRT
0.955602219889006
Interpretation
Uncertain
