Gene
KWMTBOMO10909
Pre Gene Modal
BGIBMGA008484
Annotation
putative_inositol_1?3?4-triphosphate_5/6_kinase_[Danaus_plexippus]
Full name
Inositol-tetrakisphosphate 1-kinase
Location in the cell
Cytoplasmic Reliability : 1.34 Nuclear Reliability : 1.008 PlasmaMembrane Reliability : 1.618
Sequence
CDS
ATGGAAAGTAACTCGTTTAATGAAACGATCGGTATATGGATATCCGACAAAAAGAGCCAAAAACTTAATTGGAAAGAATTAATTAATACATGCGCTGCGTTTAACTATAATGTTGTCAAGTTGGATTTAGATCAATCTTTAGATGACCAAGCACGAATAGACGTGTTCCTTCACAAACTCACTGATATTATAGCAGCTGCTGATCAAGGGGATCCAAAAGCATCGATAATAATCAACCGAGTGGAACAGTTTTTATCAAATCATCCAGAAATCGTTGTCATCGATCCCTTAGAAAGTGTCAGGATTTTACTGAATAGAAATTGTTACTATTCGATACTCCAGGATGAAACGTCTTTCTCAAACCAAGGGATATTCACACCTAATTTTGCAGAGTTCACGTATAATGTAATAGAAAGGAATATTGAAATAATGAAACAGAGAGGCGTCACATTTCCATTAGTTTGCAAACCTACATTAGCGCACGGTTCGAAGTCTGCACATGAAATGGTAATAATATTTAATGAAAAGGATCTTGGTGCTTGCAAACCGCCATGTGTTGTGCAAAGTTTTGTGAACCACAATGCTGTCCTCCATAAAGTATTTTTAGTAGGAGATCGATACCACATCTGTGAGCGGCCAAGCATTAAAAATTTTACATCATCCGAAAATTTGGATCCAATATATTACAGTACCGGGGAGGTCATCAATTGGATATTTAAAGCTGGTAGGCGTACGAAGTGCTATCCCCATAACATAACCACATATATTGTCTACAGATTTTGA
Protein
MESNSFNETIGIWISDKKSQKLNWKELINTCAAFNYNVVKLDLDQSLDDQARIDVFLHKLTDIIAAADQGDPKASIIINRVEQFLSNHPEIVVIDPLESVRILLNRNCYYSILQDETSFSNQGIFTPNFAEFTYNVIERNIEIMKQRGVTFPLVCKPTLAHGSKSAHEMVIIFNEKDLGACKPPCVVQSFVNHNAVLHKVFLVGDRYHICERPSIKNFTSSENLDPIYYSTGEVINWIFKAGRRTKCYPHNITTYIVYRF
Summary
Description
Kinase that can phosphorylate various inositol polyphosphate such as Ins(3,4,5,6)P4 or Ins(1,3,4)P3.
Catalytic Activity
1D-myo-inositol 3,4,5,6-tetrakisphosphate + ATP = 1D-myo-inositol 1,3,4,5,6-pentakisphosphate + ADP + H(+)
Cofactor
Mg(2+)
Subunit
Monomer.
Similarity
Belongs to the ITPK1 family.
Feature
chain Inositol-tetrakisphosphate 1-kinase
Uniprot
H9JG37
A0A2W1BPU4
A0A2H1VBL6
A0A212F8P9
A0A1E1WJ36
A0A194QME8
+ More
A0A194PGR7 A0A2J7PEI4 A0A2J7PEI6 D2A1U2 T1HI46 A0A1W4W6D3 A0A2S2Q6A0 A0A023F3E0 A0A2J7PSR2 A0A0P4VNS0 A0A2H8THT3 A0A069DSJ4 A0A1B6MHQ6 A0A2R7W2S8 A0A067QWY4 A0A1B6G3F3 J9JK17 A0A1B6HI43 A0A1B6HVI2 E1ZWB4 A0A1Y1L2D3 A0A1Y1L2P2 F4WX36 A0A154PJ25 A0A158NCI3 A0A0A9Y7G0 A0A195BXD4 A0A195ELL6 A0A0K8SQ66 A0A195FAD1 E2BBW2 A0A3L8DHQ0 A0A026X179 N6TRB2 J3JU34 A0A151X0F6 A0A151IQT5 A0A2J7PEI7 H2T0A5 A0A088AAM3 H3DJ06 A0A3P8UHF1 K7J9R5 A0A3Q0T5G9 A0A0S7GTR7 A0A3Q3X045 A0A3Q1AZJ1 A0A3B5ADP3 A0A1A8N7L4 A0A1A8N2H6 A0A1A8E7R8 A0A1A8A1T3 A0A3B3HPP4 A0A3P9M0E3 A0A3Q1JFK6 A0A1A8H7Q9 I3KW72 A0A0T6B184 A0A3Q1JMA6 A0A0S7GTU5 A0A3Q4GM20 A0A2I4BN11 A0A3P8UHD7 A0A3Q1JFN4 A0A0F8C6R1 A0A2I4BMX4 A0A3B4X019 A0A3B4T6Q8 A0A3B3D4P3 A0A3Q3DN98 G3NZR8 A0A2I4BMW6 A0A3Q3GB12 A0A3Q3JDI0 A0A1B6CW31 A0A3Q3L3Z6 A0A3B4T6I8 A0A3B4X000 A0A0S7GWK4 A0A3P8QSY7 A0A3P9BLT3 A0A1V4KAX8 A0A1U7SLW0 A0A0L7QP61 A0A3B3YY89 A0A3B4H9I9 A0A3Q3A6S8 A0A0S7GTP6 A0A1D5NV90 A0A226ESF8 A0A3B4A6T4 M4AGL1 A0A3P9PIG6 G1KI84
A0A194PGR7 A0A2J7PEI4 A0A2J7PEI6 D2A1U2 T1HI46 A0A1W4W6D3 A0A2S2Q6A0 A0A023F3E0 A0A2J7PSR2 A0A0P4VNS0 A0A2H8THT3 A0A069DSJ4 A0A1B6MHQ6 A0A2R7W2S8 A0A067QWY4 A0A1B6G3F3 J9JK17 A0A1B6HI43 A0A1B6HVI2 E1ZWB4 A0A1Y1L2D3 A0A1Y1L2P2 F4WX36 A0A154PJ25 A0A158NCI3 A0A0A9Y7G0 A0A195BXD4 A0A195ELL6 A0A0K8SQ66 A0A195FAD1 E2BBW2 A0A3L8DHQ0 A0A026X179 N6TRB2 J3JU34 A0A151X0F6 A0A151IQT5 A0A2J7PEI7 H2T0A5 A0A088AAM3 H3DJ06 A0A3P8UHF1 K7J9R5 A0A3Q0T5G9 A0A0S7GTR7 A0A3Q3X045 A0A3Q1AZJ1 A0A3B5ADP3 A0A1A8N7L4 A0A1A8N2H6 A0A1A8E7R8 A0A1A8A1T3 A0A3B3HPP4 A0A3P9M0E3 A0A3Q1JFK6 A0A1A8H7Q9 I3KW72 A0A0T6B184 A0A3Q1JMA6 A0A0S7GTU5 A0A3Q4GM20 A0A2I4BN11 A0A3P8UHD7 A0A3Q1JFN4 A0A0F8C6R1 A0A2I4BMX4 A0A3B4X019 A0A3B4T6Q8 A0A3B3D4P3 A0A3Q3DN98 G3NZR8 A0A2I4BMW6 A0A3Q3GB12 A0A3Q3JDI0 A0A1B6CW31 A0A3Q3L3Z6 A0A3B4T6I8 A0A3B4X000 A0A0S7GWK4 A0A3P8QSY7 A0A3P9BLT3 A0A1V4KAX8 A0A1U7SLW0 A0A0L7QP61 A0A3B3YY89 A0A3B4H9I9 A0A3Q3A6S8 A0A0S7GTP6 A0A1D5NV90 A0A226ESF8 A0A3B4A6T4 M4AGL1 A0A3P9PIG6 G1KI84
EC Number
2.7.1.134
Pubmed
EMBL
BABH01001884
BABH01001885
BABH01001886
KZ149999
PZC75337.1
ODYU01001676
+ More
SOQ38229.1 AGBW02009692 OWR50107.1 GDQN01004035 JAT87019.1 KQ461196 KPJ06534.1 KQ459604 KPI92581.1 NEVH01026094 PNF14750.1 PNF14751.1 KQ971338 EFA02124.1 ACPB03008780 GGMS01003927 MBY73130.1 GBBI01002899 JAC15813.1 NEVH01021924 PNF19372.1 GDKW01000258 JAI56337.1 GFXV01001775 MBW13580.1 GBGD01001831 JAC87058.1 GEBQ01004524 JAT35453.1 KK854263 PTY13899.1 KK853150 KDR10607.1 GECZ01012799 JAS56970.1 ABLF02020150 ABLF02020152 GECU01036116 GECU01033374 JAS71590.1 JAS74332.1 GECU01029071 GECU01022143 JAS78635.1 JAS85563.1 GL434791 EFN74518.1 GEZM01066507 JAV67843.1 GEZM01066506 JAV67844.1 GL888417 EGI61279.1 KQ434908 KZC11308.1 ADTU01011907 ADTU01011908 GBHO01018119 GDHC01015325 JAG25485.1 JAQ03304.1 KQ976396 KYM92955.1 KQ978691 KYN29170.1 GBRD01010874 JAG54950.1 KQ981727 KYN37004.1 GL447175 EFN86786.1 QOIP01000008 RLU19722.1 KK107034 EZA62042.1 APGK01057768 APGK01057769 APGK01057770 KB741281 ENN70861.1 BT126746 AEE61708.1 KQ982614 KYQ53834.1 KQ976759 KYN08554.1 PNF14753.1 AAZX01001623 AAZX01014080 GBYX01448959 JAO32524.1 HAEH01000416 HAEI01006639 SBR64847.1 HAEF01003131 HAEG01000111 SBR63315.1 HADZ01006128 HAEA01012664 SBQ41144.1 HADY01010607 HAEJ01011662 SBP49092.1 HAEC01012034 SBQ80251.1 AERX01030261 AERX01030262 LJIG01016261 KRT81132.1 GBYX01448949 JAO32534.1 KQ042668 KKF12127.1 GEDC01019608 JAS17690.1 GBYX01448960 JAO32523.1 LSYS01003973 OPJ81544.1 KQ414843 KOC60339.1 GBYX01448950 JAO32533.1 AADN04000016 LNIX01000002 OXA60542.1
SOQ38229.1 AGBW02009692 OWR50107.1 GDQN01004035 JAT87019.1 KQ461196 KPJ06534.1 KQ459604 KPI92581.1 NEVH01026094 PNF14750.1 PNF14751.1 KQ971338 EFA02124.1 ACPB03008780 GGMS01003927 MBY73130.1 GBBI01002899 JAC15813.1 NEVH01021924 PNF19372.1 GDKW01000258 JAI56337.1 GFXV01001775 MBW13580.1 GBGD01001831 JAC87058.1 GEBQ01004524 JAT35453.1 KK854263 PTY13899.1 KK853150 KDR10607.1 GECZ01012799 JAS56970.1 ABLF02020150 ABLF02020152 GECU01036116 GECU01033374 JAS71590.1 JAS74332.1 GECU01029071 GECU01022143 JAS78635.1 JAS85563.1 GL434791 EFN74518.1 GEZM01066507 JAV67843.1 GEZM01066506 JAV67844.1 GL888417 EGI61279.1 KQ434908 KZC11308.1 ADTU01011907 ADTU01011908 GBHO01018119 GDHC01015325 JAG25485.1 JAQ03304.1 KQ976396 KYM92955.1 KQ978691 KYN29170.1 GBRD01010874 JAG54950.1 KQ981727 KYN37004.1 GL447175 EFN86786.1 QOIP01000008 RLU19722.1 KK107034 EZA62042.1 APGK01057768 APGK01057769 APGK01057770 KB741281 ENN70861.1 BT126746 AEE61708.1 KQ982614 KYQ53834.1 KQ976759 KYN08554.1 PNF14753.1 AAZX01001623 AAZX01014080 GBYX01448959 JAO32524.1 HAEH01000416 HAEI01006639 SBR64847.1 HAEF01003131 HAEG01000111 SBR63315.1 HADZ01006128 HAEA01012664 SBQ41144.1 HADY01010607 HAEJ01011662 SBP49092.1 HAEC01012034 SBQ80251.1 AERX01030261 AERX01030262 LJIG01016261 KRT81132.1 GBYX01448949 JAO32534.1 KQ042668 KKF12127.1 GEDC01019608 JAS17690.1 GBYX01448960 JAO32523.1 LSYS01003973 OPJ81544.1 KQ414843 KOC60339.1 GBYX01448950 JAO32533.1 AADN04000016 LNIX01000002 OXA60542.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000053268
UP000235965
UP000007266
+ More
UP000015103 UP000192223 UP000027135 UP000007819 UP000000311 UP000007755 UP000076502 UP000005205 UP000078540 UP000078492 UP000078541 UP000008237 UP000279307 UP000053097 UP000019118 UP000075809 UP000078542 UP000005226 UP000005203 UP000007303 UP000265120 UP000002358 UP000261340 UP000261620 UP000257160 UP000261400 UP000001038 UP000265180 UP000265200 UP000265040 UP000005207 UP000261580 UP000192220 UP000261360 UP000261420 UP000261560 UP000264820 UP000007635 UP000261660 UP000261600 UP000261640 UP000265100 UP000265160 UP000190648 UP000189705 UP000053825 UP000261480 UP000261460 UP000264800 UP000000539 UP000198287 UP000261520 UP000002852 UP000242638 UP000001646
UP000015103 UP000192223 UP000027135 UP000007819 UP000000311 UP000007755 UP000076502 UP000005205 UP000078540 UP000078492 UP000078541 UP000008237 UP000279307 UP000053097 UP000019118 UP000075809 UP000078542 UP000005226 UP000005203 UP000007303 UP000265120 UP000002358 UP000261340 UP000261620 UP000257160 UP000261400 UP000001038 UP000265180 UP000265200 UP000265040 UP000005207 UP000261580 UP000192220 UP000261360 UP000261420 UP000261560 UP000264820 UP000007635 UP000261660 UP000261600 UP000261640 UP000265100 UP000265160 UP000190648 UP000189705 UP000053825 UP000261480 UP000261460 UP000264800 UP000000539 UP000198287 UP000261520 UP000002852 UP000242638 UP000001646
PRIDE
Interpro
ProteinModelPortal
H9JG37
A0A2W1BPU4
A0A2H1VBL6
A0A212F8P9
A0A1E1WJ36
A0A194QME8
+ More
A0A194PGR7 A0A2J7PEI4 A0A2J7PEI6 D2A1U2 T1HI46 A0A1W4W6D3 A0A2S2Q6A0 A0A023F3E0 A0A2J7PSR2 A0A0P4VNS0 A0A2H8THT3 A0A069DSJ4 A0A1B6MHQ6 A0A2R7W2S8 A0A067QWY4 A0A1B6G3F3 J9JK17 A0A1B6HI43 A0A1B6HVI2 E1ZWB4 A0A1Y1L2D3 A0A1Y1L2P2 F4WX36 A0A154PJ25 A0A158NCI3 A0A0A9Y7G0 A0A195BXD4 A0A195ELL6 A0A0K8SQ66 A0A195FAD1 E2BBW2 A0A3L8DHQ0 A0A026X179 N6TRB2 J3JU34 A0A151X0F6 A0A151IQT5 A0A2J7PEI7 H2T0A5 A0A088AAM3 H3DJ06 A0A3P8UHF1 K7J9R5 A0A3Q0T5G9 A0A0S7GTR7 A0A3Q3X045 A0A3Q1AZJ1 A0A3B5ADP3 A0A1A8N7L4 A0A1A8N2H6 A0A1A8E7R8 A0A1A8A1T3 A0A3B3HPP4 A0A3P9M0E3 A0A3Q1JFK6 A0A1A8H7Q9 I3KW72 A0A0T6B184 A0A3Q1JMA6 A0A0S7GTU5 A0A3Q4GM20 A0A2I4BN11 A0A3P8UHD7 A0A3Q1JFN4 A0A0F8C6R1 A0A2I4BMX4 A0A3B4X019 A0A3B4T6Q8 A0A3B3D4P3 A0A3Q3DN98 G3NZR8 A0A2I4BMW6 A0A3Q3GB12 A0A3Q3JDI0 A0A1B6CW31 A0A3Q3L3Z6 A0A3B4T6I8 A0A3B4X000 A0A0S7GWK4 A0A3P8QSY7 A0A3P9BLT3 A0A1V4KAX8 A0A1U7SLW0 A0A0L7QP61 A0A3B3YY89 A0A3B4H9I9 A0A3Q3A6S8 A0A0S7GTP6 A0A1D5NV90 A0A226ESF8 A0A3B4A6T4 M4AGL1 A0A3P9PIG6 G1KI84
A0A194PGR7 A0A2J7PEI4 A0A2J7PEI6 D2A1U2 T1HI46 A0A1W4W6D3 A0A2S2Q6A0 A0A023F3E0 A0A2J7PSR2 A0A0P4VNS0 A0A2H8THT3 A0A069DSJ4 A0A1B6MHQ6 A0A2R7W2S8 A0A067QWY4 A0A1B6G3F3 J9JK17 A0A1B6HI43 A0A1B6HVI2 E1ZWB4 A0A1Y1L2D3 A0A1Y1L2P2 F4WX36 A0A154PJ25 A0A158NCI3 A0A0A9Y7G0 A0A195BXD4 A0A195ELL6 A0A0K8SQ66 A0A195FAD1 E2BBW2 A0A3L8DHQ0 A0A026X179 N6TRB2 J3JU34 A0A151X0F6 A0A151IQT5 A0A2J7PEI7 H2T0A5 A0A088AAM3 H3DJ06 A0A3P8UHF1 K7J9R5 A0A3Q0T5G9 A0A0S7GTR7 A0A3Q3X045 A0A3Q1AZJ1 A0A3B5ADP3 A0A1A8N7L4 A0A1A8N2H6 A0A1A8E7R8 A0A1A8A1T3 A0A3B3HPP4 A0A3P9M0E3 A0A3Q1JFK6 A0A1A8H7Q9 I3KW72 A0A0T6B184 A0A3Q1JMA6 A0A0S7GTU5 A0A3Q4GM20 A0A2I4BN11 A0A3P8UHD7 A0A3Q1JFN4 A0A0F8C6R1 A0A2I4BMX4 A0A3B4X019 A0A3B4T6Q8 A0A3B3D4P3 A0A3Q3DN98 G3NZR8 A0A2I4BMW6 A0A3Q3GB12 A0A3Q3JDI0 A0A1B6CW31 A0A3Q3L3Z6 A0A3B4T6I8 A0A3B4X000 A0A0S7GWK4 A0A3P8QSY7 A0A3P9BLT3 A0A1V4KAX8 A0A1U7SLW0 A0A0L7QP61 A0A3B3YY89 A0A3B4H9I9 A0A3Q3A6S8 A0A0S7GTP6 A0A1D5NV90 A0A226ESF8 A0A3B4A6T4 M4AGL1 A0A3P9PIG6 G1KI84
PDB
2QB5
E-value=2.81446e-51,
Score=508
Ontologies
PATHWAY
GO
PANTHER
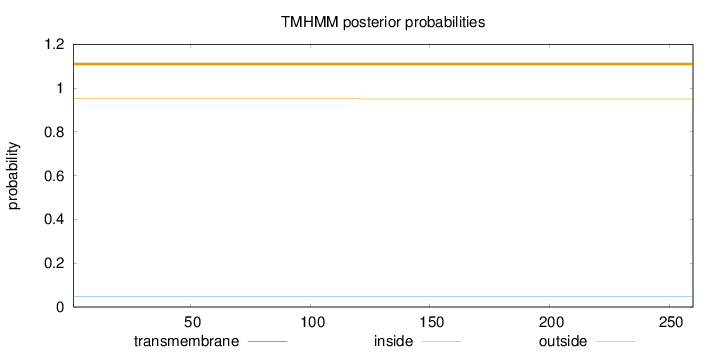
Topology
Length:
260
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00292
Exp number, first 60 AAs:
0.00037
Total prob of N-in:
0.04946
outside
1 - 260
Population Genetic Test Statistics
Pi
39.262336
Theta
15.010874
Tajima's D
-1.680709
CLR
17.208155
CSRT
0.0364481775911204
Interpretation
Uncertain
