Gene
KWMTBOMO10904 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008345
Annotation
PREDICTED:_uncharacterized_protein_LOC106719791_[Papilio_machaon]
Location in the cell
Extracellular Reliability : 2.214 Nuclear Reliability : 1.641
Sequence
CDS
ATGGCGTCATTAGCTTTGTGCTTAGTTTTATTGGCAGTTCATAATATTTTAGCAGCAGAACCCAGAAATGAAGGGTTAATGGATAAGCTGAACAATGGAATGAAATTTGCCAAAGACTTTTTGGGATCAGAATCAGTAGCTTTGAAAGTAGCAGAATTTGTAGTACGTGCATTTCAAGGTAATAAAAATCCTGAAACAAACAGAAGACACCCATTGGATGATTTGAGCAACGAAAATAATGCTTATGAAGAAAACACTGAGAAAAGTGGACCTCATTCCGAAATGAATAAAACTCCAACTCCAATGGCGCCCTTAAGACATTTAGTTAAACTTTTTGGACTTCAACCAAACCAAATAACTGCTGTGGCTGTCAATGCACTCGTTTTTGTAGCACAAATGATTTCTACTTTTCTGTCGGGTCCCAATCGTCCTTTAAAGCCGCGCTCTGACGATCCTATGACTTGGATACTGAATAAAAATTCACGTAAATTACAAGAGATGATTTCCGCTGCGAAAAATGAATCTTTACCGGACAACATCGAACATTTAATTAAACGGCAAGGTTCTGATGACGAAACGGGCTGCATTCGTCTGTTGGTTTGTAAGATTACTCCTTTTATCGTGAGGATGCAGGAGGCTGTTTTTGGTAACAACGGTCGCAACGATAAAACTAAATCGAAAGGAAATGAGCAGCAAAGAAACGGAGTAGCTTCGGTAATGTACCGCTATCTACCGACGCAAGACGAAATTAACGAACATAGCGACATCTGTGAACGACGTCACAGAGATTGTAGCATAGAATAA
Protein
MASLALCLVLLAVHNILAAEPRNEGLMDKLNNGMKFAKDFLGSESVALKVAEFVVRAFQGNKNPETNRRHPLDDLSNENNAYEENTEKSGPHSEMNKTPTPMAPLRHLVKLFGLQPNQITAVAVNALVFVAQMISTFLSGPNRPLKPRSDDPMTWILNKNSRKLQEMISAAKNESLPDNIEHLIKRQGSDDETGCIRLLVCKITPFIVRMQEAVFGNNGRNDKTKSKGNEQQRNGVASVMYRYLPTQDEINEHSDICERRHRDCSIE
Summary
Uniprot
A0A2H1X193
A0A385H8Z6
A0A2W1BJT5
A0A2A4J6G1
A0A194PGS2
A0A3S2N8T5
+ More
A0A212EN09 A0A194QSL8 A0A0L7KQ33 A0A0L7LKX0 A0A2J7PUI7 A0A310S733 A0A2A3ES61 A0A087ZUS1 D2A1C2 A0A0M8ZU27 E2BSW7 A0A0M4EGP4 E2AL59 A0A154P2J1 A0A1B0A752 A0A182LRK7 B4HKM9 A0A1A9WEB9 A0A3L8E230 A0A1W4VAR9 Q17BN7 B4IXV7 B3M6D8 A0A1B0BXR5 A0A195CC17 B4QMR9 E9IZT2 Q3ZAK9 A0A182WBD6 Q8IQC7 B0X015 A0A182G159 A0A182QIM9 B4LBA3 A0A1I8PUD5 A0A182N9F6 A0A151WV47 A0A182SDV8 B4N6Y0 A0A195FDH5 A0A182I6L2 B3NCD7 A0A182RJF8 A0A1A9V5Y8 A0A0A1WZ33 A0A3B0KJU5 B4KVE7 A0A182WWD3 A0A182V1B1 A0A182L3T4 B4PEZ5 A0A182PIN7 A0A182TIV4 A0A182J3C1 A8E6P7 A0A026W5J7 A0A1A9XD85 A0A0L0BXJ1 B4HA65 Q29F73 A0A182Y823 A0A2S2P811 T1IMM7
A0A212EN09 A0A194QSL8 A0A0L7KQ33 A0A0L7LKX0 A0A2J7PUI7 A0A310S733 A0A2A3ES61 A0A087ZUS1 D2A1C2 A0A0M8ZU27 E2BSW7 A0A0M4EGP4 E2AL59 A0A154P2J1 A0A1B0A752 A0A182LRK7 B4HKM9 A0A1A9WEB9 A0A3L8E230 A0A1W4VAR9 Q17BN7 B4IXV7 B3M6D8 A0A1B0BXR5 A0A195CC17 B4QMR9 E9IZT2 Q3ZAK9 A0A182WBD6 Q8IQC7 B0X015 A0A182G159 A0A182QIM9 B4LBA3 A0A1I8PUD5 A0A182N9F6 A0A151WV47 A0A182SDV8 B4N6Y0 A0A195FDH5 A0A182I6L2 B3NCD7 A0A182RJF8 A0A1A9V5Y8 A0A0A1WZ33 A0A3B0KJU5 B4KVE7 A0A182WWD3 A0A182V1B1 A0A182L3T4 B4PEZ5 A0A182PIN7 A0A182TIV4 A0A182J3C1 A8E6P7 A0A026W5J7 A0A1A9XD85 A0A0L0BXJ1 B4HA65 Q29F73 A0A182Y823 A0A2S2P811 T1IMM7
Pubmed
EMBL
ODYU01012210
SOQ58394.1
MH028122
AXY04291.1
KZ149999
PZC75342.1
+ More
NWSH01002670 PCG67707.1 KQ459604 KPI92586.1 RSAL01000195 RVE44568.1 AGBW02013764 OWR42866.1 KQ461196 KPJ06536.1 JTDY01007501 KOB65156.1 JTDY01000751 KOB76014.1 NEVH01021197 PNF19997.1 KQ766789 OAD53567.1 KZ288193 PBC34112.1 KQ971338 EFA01551.1 KQ435845 KOX71170.1 GL450306 EFN81215.1 CP012525 ALC43627.1 GL440492 EFN65814.1 KQ434804 KZC06052.1 AXCM01021014 CH480815 EDW40832.1 QOIP01000001 RLU26780.1 CH477319 EAT43676.1 CH916366 EDV96478.1 CH902618 EDV40787.1 JXJN01022301 KQ978068 KYM97628.1 CM000363 EDX09823.1 GL767232 EFZ13946.1 BT023860 AAZ86781.1 AE014296 AAN11958.1 DS232225 EDS37890.1 JXUM01136077 KQ568375 KXJ69025.1 AXCN02000560 CH940647 EDW69691.1 KQ982730 KYQ51525.1 CH964168 EDW80119.1 KQ981677 KYN38099.1 APCN01003661 CH954178 EDV51167.1 GBXI01010614 JAD03678.1 OUUW01000009 SPP85411.1 CH933809 EDW18390.1 CM000159 EDW93050.1 BT030839 ABV82221.1 KK107432 EZA50886.1 JRES01001180 KNC24720.1 CH479237 EDW36749.1 CH379068 EAL31199.1 GGMR01012972 MBY25591.1 JH431071
NWSH01002670 PCG67707.1 KQ459604 KPI92586.1 RSAL01000195 RVE44568.1 AGBW02013764 OWR42866.1 KQ461196 KPJ06536.1 JTDY01007501 KOB65156.1 JTDY01000751 KOB76014.1 NEVH01021197 PNF19997.1 KQ766789 OAD53567.1 KZ288193 PBC34112.1 KQ971338 EFA01551.1 KQ435845 KOX71170.1 GL450306 EFN81215.1 CP012525 ALC43627.1 GL440492 EFN65814.1 KQ434804 KZC06052.1 AXCM01021014 CH480815 EDW40832.1 QOIP01000001 RLU26780.1 CH477319 EAT43676.1 CH916366 EDV96478.1 CH902618 EDV40787.1 JXJN01022301 KQ978068 KYM97628.1 CM000363 EDX09823.1 GL767232 EFZ13946.1 BT023860 AAZ86781.1 AE014296 AAN11958.1 DS232225 EDS37890.1 JXUM01136077 KQ568375 KXJ69025.1 AXCN02000560 CH940647 EDW69691.1 KQ982730 KYQ51525.1 CH964168 EDW80119.1 KQ981677 KYN38099.1 APCN01003661 CH954178 EDV51167.1 GBXI01010614 JAD03678.1 OUUW01000009 SPP85411.1 CH933809 EDW18390.1 CM000159 EDW93050.1 BT030839 ABV82221.1 KK107432 EZA50886.1 JRES01001180 KNC24720.1 CH479237 EDW36749.1 CH379068 EAL31199.1 GGMR01012972 MBY25591.1 JH431071
Proteomes
UP000218220
UP000053268
UP000283053
UP000007151
UP000053240
UP000037510
+ More
UP000235965 UP000242457 UP000005203 UP000007266 UP000053105 UP000008237 UP000092553 UP000000311 UP000076502 UP000092445 UP000075883 UP000001292 UP000091820 UP000279307 UP000192221 UP000008820 UP000001070 UP000007801 UP000092460 UP000078542 UP000000304 UP000075920 UP000000803 UP000002320 UP000069940 UP000249989 UP000075886 UP000008792 UP000095300 UP000075884 UP000075809 UP000075901 UP000007798 UP000078541 UP000075840 UP000008711 UP000075900 UP000078200 UP000268350 UP000009192 UP000076407 UP000075903 UP000075882 UP000002282 UP000075885 UP000075902 UP000075880 UP000053097 UP000092443 UP000037069 UP000008744 UP000001819 UP000076408
UP000235965 UP000242457 UP000005203 UP000007266 UP000053105 UP000008237 UP000092553 UP000000311 UP000076502 UP000092445 UP000075883 UP000001292 UP000091820 UP000279307 UP000192221 UP000008820 UP000001070 UP000007801 UP000092460 UP000078542 UP000000304 UP000075920 UP000000803 UP000002320 UP000069940 UP000249989 UP000075886 UP000008792 UP000095300 UP000075884 UP000075809 UP000075901 UP000007798 UP000078541 UP000075840 UP000008711 UP000075900 UP000078200 UP000268350 UP000009192 UP000076407 UP000075903 UP000075882 UP000002282 UP000075885 UP000075902 UP000075880 UP000053097 UP000092443 UP000037069 UP000008744 UP000001819 UP000076408
Pfam
PF09750 DRY_EERY
ProteinModelPortal
A0A2H1X193
A0A385H8Z6
A0A2W1BJT5
A0A2A4J6G1
A0A194PGS2
A0A3S2N8T5
+ More
A0A212EN09 A0A194QSL8 A0A0L7KQ33 A0A0L7LKX0 A0A2J7PUI7 A0A310S733 A0A2A3ES61 A0A087ZUS1 D2A1C2 A0A0M8ZU27 E2BSW7 A0A0M4EGP4 E2AL59 A0A154P2J1 A0A1B0A752 A0A182LRK7 B4HKM9 A0A1A9WEB9 A0A3L8E230 A0A1W4VAR9 Q17BN7 B4IXV7 B3M6D8 A0A1B0BXR5 A0A195CC17 B4QMR9 E9IZT2 Q3ZAK9 A0A182WBD6 Q8IQC7 B0X015 A0A182G159 A0A182QIM9 B4LBA3 A0A1I8PUD5 A0A182N9F6 A0A151WV47 A0A182SDV8 B4N6Y0 A0A195FDH5 A0A182I6L2 B3NCD7 A0A182RJF8 A0A1A9V5Y8 A0A0A1WZ33 A0A3B0KJU5 B4KVE7 A0A182WWD3 A0A182V1B1 A0A182L3T4 B4PEZ5 A0A182PIN7 A0A182TIV4 A0A182J3C1 A8E6P7 A0A026W5J7 A0A1A9XD85 A0A0L0BXJ1 B4HA65 Q29F73 A0A182Y823 A0A2S2P811 T1IMM7
A0A212EN09 A0A194QSL8 A0A0L7KQ33 A0A0L7LKX0 A0A2J7PUI7 A0A310S733 A0A2A3ES61 A0A087ZUS1 D2A1C2 A0A0M8ZU27 E2BSW7 A0A0M4EGP4 E2AL59 A0A154P2J1 A0A1B0A752 A0A182LRK7 B4HKM9 A0A1A9WEB9 A0A3L8E230 A0A1W4VAR9 Q17BN7 B4IXV7 B3M6D8 A0A1B0BXR5 A0A195CC17 B4QMR9 E9IZT2 Q3ZAK9 A0A182WBD6 Q8IQC7 B0X015 A0A182G159 A0A182QIM9 B4LBA3 A0A1I8PUD5 A0A182N9F6 A0A151WV47 A0A182SDV8 B4N6Y0 A0A195FDH5 A0A182I6L2 B3NCD7 A0A182RJF8 A0A1A9V5Y8 A0A0A1WZ33 A0A3B0KJU5 B4KVE7 A0A182WWD3 A0A182V1B1 A0A182L3T4 B4PEZ5 A0A182PIN7 A0A182TIV4 A0A182J3C1 A8E6P7 A0A026W5J7 A0A1A9XD85 A0A0L0BXJ1 B4HA65 Q29F73 A0A182Y823 A0A2S2P811 T1IMM7
Ontologies
GO
PANTHER
Topology
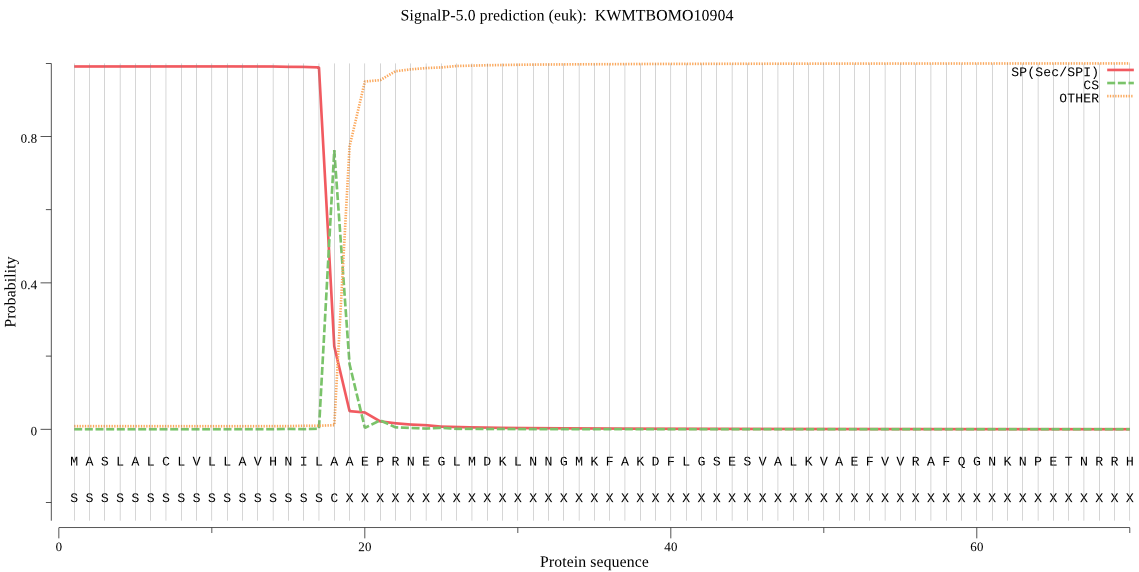
SignalP
Position: 1 - 18,
Likelihood: 0.991180
Length:
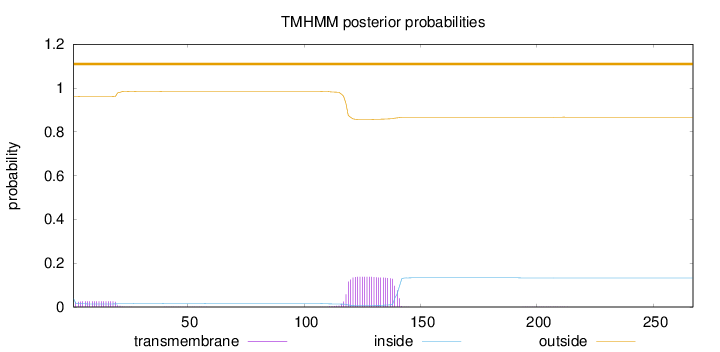
267
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.49163
Exp number, first 60 AAs:
0.47283
Total prob of N-in:
0.03883
outside
1 - 267
Population Genetic Test Statistics
Pi
169.716046
Theta
151.139825
Tajima's D
0.479578
CLR
0.1034
CSRT
0.509024548772561
Interpretation
Possibly Positive selection
