Pre Gene Modal
BGIBMGA008478
Annotation
PREDICTED:_ribokinase-like_isoform_X2_[Bombyx_mori]
Full name
Ribokinase
Location in the cell
Cytoplasmic Reliability : 1.809 Extracellular Reliability : 1.383
Sequence
CDS
ATGGATACCAAAAATGGTTATAAACTTGTAGTTCTTGGCTCTTGTAGTGTGGATTTTACAACATATGCACCGAAACTGCCTAAATCTGGAGAAACTTTACATGGTACAAAATTCACTACCAGTTTCGGAGGAAAAGGTGCAAACCAGTGTGTTGCTGCTGCGAAATTAGGTGGAAATGCCTACATGATTTGTAGAGTGGGGGATGATCAGTGGGGTAAAAAATATAAAGACCATCTCAAAAATGAAGGTGTTAATGTGTCTTATGTTCACATCACAAAGAATGAAACAACAGGCGTAGCACAAATTGTTGTAGCAGAAAACGGAGAAAATCAAATCGTTATTGTACCGGGAGCTAATAAATGTCTAAGTGTCCAAGATGTAGAAGAGTCAATTGAATTGATAAAAAATGCTGATGTTCTAATAGGACAGCTTGAAACTCCGTTTGAAACTACTTATACAGCATTTAAGTTAAATAATGGTATAAAACTATTAAATGCCGCTCCAGCGTTGACTGATATTCGGAAAATTTTGCCATTTTGTACTATACTTTGTGTTAATGAATTAGAAGCATCAGTTCTCACTAATGTGGATGTCACCATTTCGAATGCTTCAATGGCTGCAAAAAAATTGCTGGAAACTGGCTGCGAAACAGTTATAATAACACTAGGATCTGAAGGAGCTGTTTATATGTCAAAAAACGAAGAATGTCCTAAACATGTTTTATGTGAAGCTGTTAATCCTGTAGATACCACAGGTGCCGGCGATGCTTTCATTGGAGCTTTGGCAACATTTCTGGTCGAGTATAAAGATTATCCACTGCACCAATTAATCGGCGCAGCTTGTGCGGTCGCAACCATTTCAGTTACAGTGGAAGGCACACAAACGAGTTATCCTACGAAAAATGATTTCTTTTGTAAAAAGTATGAGTACGTCCAACTGGAATAA
Protein
MDTKNGYKLVVLGSCSVDFTTYAPKLPKSGETLHGTKFTTSFGGKGANQCVAAAKLGGNAYMICRVGDDQWGKKYKDHLKNEGVNVSYVHITKNETTGVAQIVVAENGENQIVIVPGANKCLSVQDVEESIELIKNADVLIGQLETPFETTYTAFKLNNGIKLLNAAPALTDIRKILPFCTILCVNELEASVLTNVDVTISNASMAAKKLLETGCETVIITLGSEGAVYMSKNEECPKHVLCEAVNPVDTTGAGDAFIGALATFLVEYKDYPLHQLIGAACAVATISVTVEGTQTSYPTKNDFFCKKYEYVQLE
Summary
Description
Catalyzes the phosphorylation of ribose at O-5 in a reaction requiring ATP and magnesium. The resulting D-ribose-5-phosphate can then be used either for sythesis of nucleotides, histidine, and tryptophan, or as a component of the pentose phosphate pathway.
Catalytic Activity
ATP + D-ribose = ADP + D-ribose 5-phosphate + H(+)
Cofactor
Mg(2+)
Subunit
Homodimer.
Similarity
Belongs to the carbohydrate kinase PfkB family. Ribokinase subfamily.
Belongs to the carbohydrate kinase PfkB family.
Belongs to the carbohydrate kinase PfkB family.
Uniprot
A0A2H1WTD3
A0A2A4J865
A0A2W1BPV4
A0A212EN32
A0A3S2LDV1
A0A194PGS7
+ More
S4P727 A0A194QMG2 A0A220K8M3 U5EU95 A0A182MQH6 A0A2M4BTY5 A0A182W0V7 W5JR37 A0A0A1X5S4 A0A182FT02 A0A182Y6J7 D6WXB9 A0A034WHI3 B0X0B9 A0A182INJ9 A0A1I8NU87 Q17H56 A0A1W4WDZ1 A0A182PZV3 A0A1Q3FKN1 A0A0K8UGQ9 W8BYA7 A0A084WTS6 A0A2M4BUZ1 A0A0A1XFH1 A0A182RLD8 A0A1L8ECJ5 A0A1L8ECS4 A0A023EPD7 A0A154P6D5 R4WIW3 A0A1L8EBZ7 A0A1L8E1X0 A0A232FKH2 A0A182PCC4 A0A1L8E1M4 A0A182VEN7 A0A023EPF3 A0A182NRG7 A0A1I8MI95 K7J480 A0A0P4VXF9 A0A0K8VLV4 A0A158NGU0 A0A182UIL9 F4WZP8 W8CBB7 E9IUQ2 A0A182HSJ4 A0A0L0C667 A0A0K8TMI2 Q7QCF9 A0A182XAQ4 B4JK94 A0A1B6BYR2 A0A026W9W0 B3P982 E2BVL2 A0A0A9WXY4 E2AJB5 A0A1Y1M6B7 A0A0K8SLC0 B4PX14 A0A1W7R8J4 A0A3M6T4E6 O77425 A0A3B0JDK0 Q29I50 A0A2B4RFF0 B3MYB0 B4I938 R4FND1 B4M9X8 A0A0M5JE21 A0A034WK55 A0A2A3ECU7 L8I2X8 B4NEA9 A0A088ARF3 A0A1U7UQC7 A0A2Y9HNP7 A0A182JYQ6 M3W036 W5QDU7 A0A3P9A491 G1KWF4 G1SZT1 A0A1B0AFE2 A0A369SKR6 A0A0L7R4A1 A0A0P4VJH2 A0A151XCW2 A0A1B0GQU7 A0A3Q7NR74
S4P727 A0A194QMG2 A0A220K8M3 U5EU95 A0A182MQH6 A0A2M4BTY5 A0A182W0V7 W5JR37 A0A0A1X5S4 A0A182FT02 A0A182Y6J7 D6WXB9 A0A034WHI3 B0X0B9 A0A182INJ9 A0A1I8NU87 Q17H56 A0A1W4WDZ1 A0A182PZV3 A0A1Q3FKN1 A0A0K8UGQ9 W8BYA7 A0A084WTS6 A0A2M4BUZ1 A0A0A1XFH1 A0A182RLD8 A0A1L8ECJ5 A0A1L8ECS4 A0A023EPD7 A0A154P6D5 R4WIW3 A0A1L8EBZ7 A0A1L8E1X0 A0A232FKH2 A0A182PCC4 A0A1L8E1M4 A0A182VEN7 A0A023EPF3 A0A182NRG7 A0A1I8MI95 K7J480 A0A0P4VXF9 A0A0K8VLV4 A0A158NGU0 A0A182UIL9 F4WZP8 W8CBB7 E9IUQ2 A0A182HSJ4 A0A0L0C667 A0A0K8TMI2 Q7QCF9 A0A182XAQ4 B4JK94 A0A1B6BYR2 A0A026W9W0 B3P982 E2BVL2 A0A0A9WXY4 E2AJB5 A0A1Y1M6B7 A0A0K8SLC0 B4PX14 A0A1W7R8J4 A0A3M6T4E6 O77425 A0A3B0JDK0 Q29I50 A0A2B4RFF0 B3MYB0 B4I938 R4FND1 B4M9X8 A0A0M5JE21 A0A034WK55 A0A2A3ECU7 L8I2X8 B4NEA9 A0A088ARF3 A0A1U7UQC7 A0A2Y9HNP7 A0A182JYQ6 M3W036 W5QDU7 A0A3P9A491 G1KWF4 G1SZT1 A0A1B0AFE2 A0A369SKR6 A0A0L7R4A1 A0A0P4VJH2 A0A151XCW2 A0A1B0GQU7 A0A3Q7NR74
EC Number
2.7.1.15
Pubmed
28756777
22118469
26354079
23622113
28630352
20920257
+ More
23761445 25830018 25244985 18362917 19820115 25348373 17510324 24495485 24438588 24945155 23691247 28648823 25315136 20075255 21347285 21719571 21282665 26108605 26369729 12364791 14747013 17210077 17994087 24508170 20798317 25401762 26823975 28004739 17550304 30382153 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 22751099 17975172 20809919 25069045 21993624 30042472 27129103
23761445 25830018 25244985 18362917 19820115 25348373 17510324 24495485 24438588 24945155 23691247 28648823 25315136 20075255 21347285 21719571 21282665 26108605 26369729 12364791 14747013 17210077 17994087 24508170 20798317 25401762 26823975 28004739 17550304 30382153 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 22751099 17975172 20809919 25069045 21993624 30042472 27129103
EMBL
ODYU01010526
SOQ55694.1
NWSH01002670
PCG67714.1
KZ149999
PZC75347.1
+ More
AGBW02013764 OWR42861.1 RSAL01000195 RVE44565.1 KQ459604 KPI92591.1 GAIX01004779 JAA87781.1 KQ461196 KPJ06544.1 MF319724 ASJ26438.1 GANO01002422 JAB57449.1 AXCM01000145 GGFJ01007396 MBW56537.1 ADMH02000418 ETN66586.1 GBXI01008269 JAD06023.1 KQ971361 EFA08820.2 GAKP01004803 JAC54149.1 DS232232 EDS38080.1 CH477253 EAT45957.1 AXCN02001460 GFDL01006908 JAV28137.1 GDHF01030759 GDHF01026465 JAI21555.1 JAI25849.1 GAMC01004962 JAC01594.1 ATLV01026914 KE525420 KFB53620.1 GGFJ01007692 MBW56833.1 GBXI01004612 JAD09680.1 GFDG01002349 JAV16450.1 GFDG01002353 JAV16446.1 GAPW01002737 JAC10861.1 KQ434826 KZC07401.1 AK417425 BAN20640.1 GFDG01002654 JAV16145.1 GFDF01001455 JAV12629.1 NNAY01000080 OXU31165.1 GFDF01001454 JAV12630.1 GAPW01002717 JAC10881.1 AAZX01010390 GDRN01089668 GDRN01089666 JAI60615.1 GDHF01012452 JAI39862.1 ADTU01015211 GL888480 EGI60323.1 GAMC01004961 JAC01595.1 GL766012 EFZ15684.1 APCN01000281 JRES01000948 KNC26904.1 GDAI01002039 JAI15564.1 AAAB01008859 EAA08069.5 CH916370 EDV99996.1 GEDC01030886 GEDC01018566 JAS06412.1 JAS18732.1 KK107314 EZA52835.1 CH954183 EDV45378.1 GL450875 EFN80282.1 GBHO01032211 GBHO01018892 GDHC01002807 JAG11393.1 JAG24712.1 JAQ15822.1 GL439972 EFN66436.1 GEZM01042455 JAV79845.1 GBRD01011713 JAG54111.1 CM000162 EDX00800.1 GEHC01000192 JAV47453.1 RCHS01004356 RMX35519.1 AE014298 BT021233 AL031581 AAF45527.1 AAX33381.1 CAA20884.1 OUUW01000003 SPP78212.1 CH379064 EAL31556.2 LSMT01000497 PFX17094.1 CH902632 EDV32604.1 CH480825 EDW43719.1 ACPB03015647 GAHY01001344 JAA76166.1 CH940655 EDW66037.1 CP012528 ALC49981.1 GAKP01004805 GAKP01004804 JAC54148.1 KZ288282 PBC29603.1 JH882185 ELR50466.1 CH964239 EDW82078.1 AANG04003329 AMGL01075890 AMGL01075891 AMGL01075892 AMGL01075893 AMGL01075894 AMGL01075895 AAGW02005705 AAGW02005706 AAGW02005707 AAGW02005708 AAGW02005709 NOWV01000003 RDD47485.1 KQ414657 KOC65678.1 GDKW01001444 JAI55151.1 KQ982294 KYQ58214.1 AJVK01019348
AGBW02013764 OWR42861.1 RSAL01000195 RVE44565.1 KQ459604 KPI92591.1 GAIX01004779 JAA87781.1 KQ461196 KPJ06544.1 MF319724 ASJ26438.1 GANO01002422 JAB57449.1 AXCM01000145 GGFJ01007396 MBW56537.1 ADMH02000418 ETN66586.1 GBXI01008269 JAD06023.1 KQ971361 EFA08820.2 GAKP01004803 JAC54149.1 DS232232 EDS38080.1 CH477253 EAT45957.1 AXCN02001460 GFDL01006908 JAV28137.1 GDHF01030759 GDHF01026465 JAI21555.1 JAI25849.1 GAMC01004962 JAC01594.1 ATLV01026914 KE525420 KFB53620.1 GGFJ01007692 MBW56833.1 GBXI01004612 JAD09680.1 GFDG01002349 JAV16450.1 GFDG01002353 JAV16446.1 GAPW01002737 JAC10861.1 KQ434826 KZC07401.1 AK417425 BAN20640.1 GFDG01002654 JAV16145.1 GFDF01001455 JAV12629.1 NNAY01000080 OXU31165.1 GFDF01001454 JAV12630.1 GAPW01002717 JAC10881.1 AAZX01010390 GDRN01089668 GDRN01089666 JAI60615.1 GDHF01012452 JAI39862.1 ADTU01015211 GL888480 EGI60323.1 GAMC01004961 JAC01595.1 GL766012 EFZ15684.1 APCN01000281 JRES01000948 KNC26904.1 GDAI01002039 JAI15564.1 AAAB01008859 EAA08069.5 CH916370 EDV99996.1 GEDC01030886 GEDC01018566 JAS06412.1 JAS18732.1 KK107314 EZA52835.1 CH954183 EDV45378.1 GL450875 EFN80282.1 GBHO01032211 GBHO01018892 GDHC01002807 JAG11393.1 JAG24712.1 JAQ15822.1 GL439972 EFN66436.1 GEZM01042455 JAV79845.1 GBRD01011713 JAG54111.1 CM000162 EDX00800.1 GEHC01000192 JAV47453.1 RCHS01004356 RMX35519.1 AE014298 BT021233 AL031581 AAF45527.1 AAX33381.1 CAA20884.1 OUUW01000003 SPP78212.1 CH379064 EAL31556.2 LSMT01000497 PFX17094.1 CH902632 EDV32604.1 CH480825 EDW43719.1 ACPB03015647 GAHY01001344 JAA76166.1 CH940655 EDW66037.1 CP012528 ALC49981.1 GAKP01004805 GAKP01004804 JAC54148.1 KZ288282 PBC29603.1 JH882185 ELR50466.1 CH964239 EDW82078.1 AANG04003329 AMGL01075890 AMGL01075891 AMGL01075892 AMGL01075893 AMGL01075894 AMGL01075895 AAGW02005705 AAGW02005706 AAGW02005707 AAGW02005708 AAGW02005709 NOWV01000003 RDD47485.1 KQ414657 KOC65678.1 GDKW01001444 JAI55151.1 KQ982294 KYQ58214.1 AJVK01019348
Proteomes
UP000218220
UP000007151
UP000283053
UP000053268
UP000053240
UP000075883
+ More
UP000075920 UP000000673 UP000069272 UP000076408 UP000007266 UP000002320 UP000075880 UP000095300 UP000008820 UP000192223 UP000075886 UP000030765 UP000075900 UP000076502 UP000215335 UP000075885 UP000075903 UP000075884 UP000095301 UP000002358 UP000005205 UP000075902 UP000007755 UP000075840 UP000037069 UP000007062 UP000076407 UP000001070 UP000053097 UP000008711 UP000008237 UP000000311 UP000002282 UP000275408 UP000000803 UP000268350 UP000001819 UP000225706 UP000007801 UP000001292 UP000015103 UP000008792 UP000092553 UP000242457 UP000007798 UP000005203 UP000189704 UP000248481 UP000075881 UP000011712 UP000002356 UP000265140 UP000001646 UP000001811 UP000092445 UP000253843 UP000053825 UP000075809 UP000092462 UP000286641
UP000075920 UP000000673 UP000069272 UP000076408 UP000007266 UP000002320 UP000075880 UP000095300 UP000008820 UP000192223 UP000075886 UP000030765 UP000075900 UP000076502 UP000215335 UP000075885 UP000075903 UP000075884 UP000095301 UP000002358 UP000005205 UP000075902 UP000007755 UP000075840 UP000037069 UP000007062 UP000076407 UP000001070 UP000053097 UP000008711 UP000008237 UP000000311 UP000002282 UP000275408 UP000000803 UP000268350 UP000001819 UP000225706 UP000007801 UP000001292 UP000015103 UP000008792 UP000092553 UP000242457 UP000007798 UP000005203 UP000189704 UP000248481 UP000075881 UP000011712 UP000002356 UP000265140 UP000001646 UP000001811 UP000092445 UP000253843 UP000053825 UP000075809 UP000092462 UP000286641
PRIDE
Pfam
PF00294 PfkB
Interpro
SUPFAM
SSF53613
SSF53613
Gene 3D
ProteinModelPortal
A0A2H1WTD3
A0A2A4J865
A0A2W1BPV4
A0A212EN32
A0A3S2LDV1
A0A194PGS7
+ More
S4P727 A0A194QMG2 A0A220K8M3 U5EU95 A0A182MQH6 A0A2M4BTY5 A0A182W0V7 W5JR37 A0A0A1X5S4 A0A182FT02 A0A182Y6J7 D6WXB9 A0A034WHI3 B0X0B9 A0A182INJ9 A0A1I8NU87 Q17H56 A0A1W4WDZ1 A0A182PZV3 A0A1Q3FKN1 A0A0K8UGQ9 W8BYA7 A0A084WTS6 A0A2M4BUZ1 A0A0A1XFH1 A0A182RLD8 A0A1L8ECJ5 A0A1L8ECS4 A0A023EPD7 A0A154P6D5 R4WIW3 A0A1L8EBZ7 A0A1L8E1X0 A0A232FKH2 A0A182PCC4 A0A1L8E1M4 A0A182VEN7 A0A023EPF3 A0A182NRG7 A0A1I8MI95 K7J480 A0A0P4VXF9 A0A0K8VLV4 A0A158NGU0 A0A182UIL9 F4WZP8 W8CBB7 E9IUQ2 A0A182HSJ4 A0A0L0C667 A0A0K8TMI2 Q7QCF9 A0A182XAQ4 B4JK94 A0A1B6BYR2 A0A026W9W0 B3P982 E2BVL2 A0A0A9WXY4 E2AJB5 A0A1Y1M6B7 A0A0K8SLC0 B4PX14 A0A1W7R8J4 A0A3M6T4E6 O77425 A0A3B0JDK0 Q29I50 A0A2B4RFF0 B3MYB0 B4I938 R4FND1 B4M9X8 A0A0M5JE21 A0A034WK55 A0A2A3ECU7 L8I2X8 B4NEA9 A0A088ARF3 A0A1U7UQC7 A0A2Y9HNP7 A0A182JYQ6 M3W036 W5QDU7 A0A3P9A491 G1KWF4 G1SZT1 A0A1B0AFE2 A0A369SKR6 A0A0L7R4A1 A0A0P4VJH2 A0A151XCW2 A0A1B0GQU7 A0A3Q7NR74
S4P727 A0A194QMG2 A0A220K8M3 U5EU95 A0A182MQH6 A0A2M4BTY5 A0A182W0V7 W5JR37 A0A0A1X5S4 A0A182FT02 A0A182Y6J7 D6WXB9 A0A034WHI3 B0X0B9 A0A182INJ9 A0A1I8NU87 Q17H56 A0A1W4WDZ1 A0A182PZV3 A0A1Q3FKN1 A0A0K8UGQ9 W8BYA7 A0A084WTS6 A0A2M4BUZ1 A0A0A1XFH1 A0A182RLD8 A0A1L8ECJ5 A0A1L8ECS4 A0A023EPD7 A0A154P6D5 R4WIW3 A0A1L8EBZ7 A0A1L8E1X0 A0A232FKH2 A0A182PCC4 A0A1L8E1M4 A0A182VEN7 A0A023EPF3 A0A182NRG7 A0A1I8MI95 K7J480 A0A0P4VXF9 A0A0K8VLV4 A0A158NGU0 A0A182UIL9 F4WZP8 W8CBB7 E9IUQ2 A0A182HSJ4 A0A0L0C667 A0A0K8TMI2 Q7QCF9 A0A182XAQ4 B4JK94 A0A1B6BYR2 A0A026W9W0 B3P982 E2BVL2 A0A0A9WXY4 E2AJB5 A0A1Y1M6B7 A0A0K8SLC0 B4PX14 A0A1W7R8J4 A0A3M6T4E6 O77425 A0A3B0JDK0 Q29I50 A0A2B4RFF0 B3MYB0 B4I938 R4FND1 B4M9X8 A0A0M5JE21 A0A034WK55 A0A2A3ECU7 L8I2X8 B4NEA9 A0A088ARF3 A0A1U7UQC7 A0A2Y9HNP7 A0A182JYQ6 M3W036 W5QDU7 A0A3P9A491 G1KWF4 G1SZT1 A0A1B0AFE2 A0A369SKR6 A0A0L7R4A1 A0A0P4VJH2 A0A151XCW2 A0A1B0GQU7 A0A3Q7NR74
PDB
2FV7
E-value=4.49192e-56,
Score=550
Ontologies
PATHWAY
GO
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleus
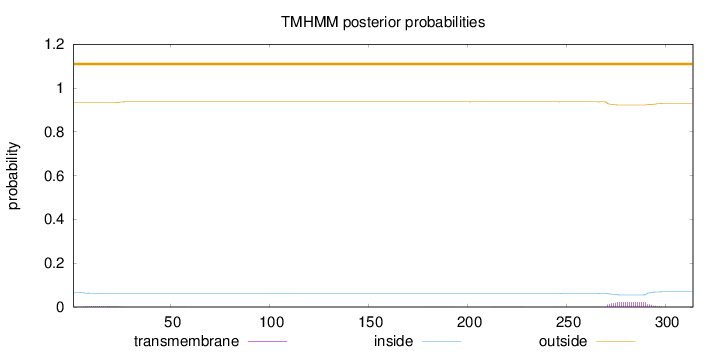
Length:
314
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.54958
Exp number, first 60 AAs:
0.05655
Total prob of N-in:
0.06602
outside
1 - 314
Population Genetic Test Statistics
Pi
375.702467
Theta
207.150625
Tajima's D
2.273843
CLR
0.24481
CSRT
0.92040397980101
Interpretation
Possibly Positive selection
