Gene
KWMTBOMO10895
Pre Gene Modal
BGIBMGA008348
Annotation
Niemann-Pick_C1_protein_[Papilio_machaon]
Location in the cell
PlasmaMembrane Reliability : 4.804
Sequence
CDS
ATGGAAGCTGTAAGTCATGTACAAGGTCTTCATGGGGAGGAAGCCGAACAAGCGATTGGAATTTTAGCCAACCGATGTCCTCATATGGTTTACGATGATGAAGGTAATATTAAACCATACGAAGAAATACTGACCTGCTGTGAGCCGATTCAAGTCATGAAGCTTTCGAGCGATTTAGCGTTGGGCGAAGGCGTTCTAGGCCGCTGTCCCACTTGCTTCAGAAACTTTGTAAGACAAATTTGCGAGATGTACTGCTCCCCTGACCAGTCTCGATTTGTGGAAGTTCATACAGAAATTGGCCCCAACAACATTGAGTATGTGGAAGCGGTTGACTACAGACTTCACGAGCCATTTATGCTGACAGCCTTCGAATCTTGCTCTGATGTTATCGTTCCTCAGACCGGTCTTCCAGCTGTTAACATGATGTGTGGCAGTGCAGTTGTATGCGATGCAGACGCGTGGTTCAAATTTACTGGCGACACTGCAGGCAACCCCATGGCACCACTTCAAATAAACTTTCAGAGATGGCCGACAGAAGAGGATTCAATGAATGTAGTTGCACTACCATGTACCGAAACTATGGAAGGTGATCTTCCGTGCAGTTGTGTTGATTGCAGTGCAATGTGTCCGGTAGGAAATAACGTTACCTATGGACCTGCATTTAGAATTGAAGCATTGCGAGAATTAGTACATCTAGAAAATAAAATCAAAAATATTGGTCGAGAACAAAGAGGTGTAACTTTAGAGGAAGTTTGCTATGCGCCTATTAGGTCCAGAGGAGCAAAGAAAGAATTAGATCAATGTGTTTATTTATCTGCATCCGTTTATTTAGGTGAAAATAGAAACGATATCGATGAAAACACTTATCTTGACGACATACAAAGCTGTTTGAACAACTTTTTTTCCTTTGACTGCTTGGCAAATTGGGGAGGAGGTGCGGAACCTGAGGCAGTTTTCGGAGGCTTTGAAGGTGATGATGTCTTGAGCGCTAATACATTGATCATTAATTTTCCTATTACCAATTTTAATTTAGAAGAAAACCTCCGTCCTGTTTTGGAATGGGAAGAGAAATTCATTGAACTTATGCAAAACTATGAAGCTTCCGGTAAATCAGATTATGTCGAAGTGGCTTTTGGAGCCGAAAGATCAATCGAAGACGAAATCAGGAGAGTTTCAGTAGCAGAAGCTGTACCGATCGCTATTAGTTATTTAATAATGTTTATTTATGTGACGTTGGCTCTAGGAAACATTAGATACTGTAGGACATGGCTCATTGATAGTAAAATATTGGTTGCTATAGGTAGTATTGTTGTTGTTGTTATCTCTATTATTTGTGCCATGGGTGTCATGGGTTATGCGAATTTCACAACTACTCTATTAGCAATAAATGTGATTCCGTTCTTCATACTTTCAGTAGGCATAGATAACGTATTCCTTATGATCAATACGTTACACGATATACAAAGTAATGTTAAAGAATACGAGGACTATGATGAGTCTTTTAGTTTTGAGAAAAAAAAGAGATTTATTTTCGAAAAAATGATGGGCAGTGTTGGCCCCTCAATGTTTGTGTCTTCTGTAACGCAAATCACCTGTTTCGCAATTGGTAGTATCACTGATGTTCCAGCGGTGGCAACATTTTCTGTATTTGCTAGCTTATCTTTGACTTTTCTTTTTATATTTCAGATAACAGCAGTTGTCGGTATACTATCATTAGACTACAAAAGAGTCGCTCAAAAACGTGTCGATATTTTTCCTTGCATTCGTAGTAAAATTTCAATGGATAGCGATCACGATCCTCCACACGTCAGTATAATGAATAGATTTATGATATTGTATTCTGATATAATTACGAATTGGAAAGTAAGAATTGTTGTTGTAATTTTCTTTATGATCATTTTATCTCTGAGTATTATTGCGGTTCCACATTTAGAAGTAGGCTTAGACCAAGAAATGGCTTTACCGACTGACTCATACGTATATAGATATTTAGTTGCTGTCAGTGAACTATTGCGTCTCGGCCCACCAGTTTATTTTGTATTAAAAGCTGGTTTAGACTTTACTAATCCGGATCACCAAAACGTTATTTGCGGAACCCAGCTGTGTTACGAAGATTCATTAATTACACAAATATTTCTAGCTGCTCAGCACAGTGAAATAACCTATATAGCAAGAAGCTCAAATTCTTGGCTGGATGACTTCTTTGATTGGTCCAGCTTACCCGGGACTTGTTGTAAATATAACGTTACCGATAATGGGTTTTGTCAAACTTTAGATACTTCTCCAGAGTGCGCCTATTGTTCGATAAGCAGGAATGAATGGTCTAACGGCCTCAGACCCAGTAAAGAGGCGTTCGAAACATTTTTACCTTTTTTCCTACAAGACGAACCAAGCCAAGTCTGCACTAAGGGAGGGTTGGCAAGTTACTCAAGCAGTGTTAATTATTTACTTGATTCTGAAGGTCGTGCCATTGTCCATGATACGAATTTTATGGCATATCAAACGGCGCTGAGCAGGTCGCCAGACTTCATCAATGCTGTGAAATATGGTTATGAGATTAGTGATAACATCACTGCCGCTATCCGGAAGCACACAGGTATGGATGTTGAAGTTTTTCCTTATTCCGTGTTTTACGTGTTTTTTGAGCAATACCTGAATATTTGGTCGGACACGTTTGCAGCGTTAGGTTTTTCAGTTTTGGGTGCTTTAGCAATAAACCTGTTAGCGTCTGGGTTTAACTTTTTGACGAGTTTCGCTGTTATATTGACTGCTGTCATGGTCGTGATCGATATGATGGGCGTAATGTATATTTGGAATATACCGCTCAATCCAGTGTCTTGCATTAATCTTATCGTGTCTATTGGTATATCGGTTGAATTTTGTAGTCATATTGCATACGCTTATGCGACAAGTCGTCGTCCAGACCACGAGAGAGTGAAAGATGCAATACAAAAAGTCGGCTCCACTGTTATTACTGGAATTACATTTACGAACATACCAATTGTAGTTTTAGCATTTTCTCACACTGAAGTTATAGAAGTGTTCTTCTTTAGGATGTTTTTTAGTCTGATAATACTAGGGTTCTTGCATGGGTTGGTGTTTTTCCCTGCTCTGCTGTTATTCCTCAATAATATAAAAACGAGATCGTTTTAA
Protein
MEAVSHVQGLHGEEAEQAIGILANRCPHMVYDDEGNIKPYEEILTCCEPIQVMKLSSDLALGEGVLGRCPTCFRNFVRQICEMYCSPDQSRFVEVHTEIGPNNIEYVEAVDYRLHEPFMLTAFESCSDVIVPQTGLPAVNMMCGSAVVCDADAWFKFTGDTAGNPMAPLQINFQRWPTEEDSMNVVALPCTETMEGDLPCSCVDCSAMCPVGNNVTYGPAFRIEALRELVHLENKIKNIGREQRGVTLEEVCYAPIRSRGAKKELDQCVYLSASVYLGENRNDIDENTYLDDIQSCLNNFFSFDCLANWGGGAEPEAVFGGFEGDDVLSANTLIINFPITNFNLEENLRPVLEWEEKFIELMQNYEASGKSDYVEVAFGAERSIEDEIRRVSVAEAVPIAISYLIMFIYVTLALGNIRYCRTWLIDSKILVAIGSIVVVVISIICAMGVMGYANFTTTLLAINVIPFFILSVGIDNVFLMINTLHDIQSNVKEYEDYDESFSFEKKKRFIFEKMMGSVGPSMFVSSVTQITCFAIGSITDVPAVATFSVFASLSLTFLFIFQITAVVGILSLDYKRVAQKRVDIFPCIRSKISMDSDHDPPHVSIMNRFMILYSDIITNWKVRIVVVIFFMIILSLSIIAVPHLEVGLDQEMALPTDSYVYRYLVAVSELLRLGPPVYFVLKAGLDFTNPDHQNVICGTQLCYEDSLITQIFLAAQHSEITYIARSSNSWLDDFFDWSSLPGTCCKYNVTDNGFCQTLDTSPECAYCSISRNEWSNGLRPSKEAFETFLPFFLQDEPSQVCTKGGLASYSSSVNYLLDSEGRAIVHDTNFMAYQTALSRSPDFINAVKYGYEISDNITAAIRKHTGMDVEVFPYSVFYVFFEQYLNIWSDTFAALGFSVLGALAINLLASGFNFLTSFAVILTAVMVVIDMMGVMYIWNIPLNPVSCINLIVSIGISVEFCSHIAYAYATSRRPDHERVKDAIQKVGSTVITGITFTNIPIVVLAFSHTEVIEVFFFRMFFSLIILGFLHGLVFFPALLLFLNNIKTRSF
Summary
Uniprot
H9JFQ1
A0A194QLW8
A0A194PNA1
A0A2W1BRF3
A0A2A4K8N4
A0A0L7L4U8
+ More
A0A1E1WQJ6 A0A2H1X3B1 A0A385NIS6 A0A2Z5U732 A0A212EN21 A0A0K8V6W5 A0A0K8WJE9 A0A0K8VI49 A0A0K8W2V5 A0A0A1WH39 A0A0A1WIT2 W8AQJ5 W8B0I9 A0A1A9UD14 A0A1B0AH79 A0A1I8M5Y1 A0A1A9Y128 A0A1B0FEI5 A0A1B0C3R6 V5GT10 V5GP54 A0A1B0C696 A0A336MXX7 A0A1A9WHG4 U4UCF2 N6TS49 B4KJH6 A0A232F5K7 D2CG00 A0A139WDV5 A0A182RV52 A0A0A9XLA8 A0A0P8Y4I3 B3MPM8 A0A182LVL1 A0A0A1XIU0 A0A182W4Q6 A0A1B0CML6 A0A182QGM2 B4L379 A0A1S4FMJ9 Q16VL3 A0A034WI75 A0A182Y7H1 A0A0K8UN34 A0A0L0C295 A0A0N0BJK7 A0A182FJQ6 A0A084VJF0 A0A1L8DK46 A0A1L8DXZ7 A0A1L8DJW6 A0A1I8PCF8 A0A1L8DK92 U5EYU3 A0A0K8S3E0 A0A0K8SKT4 N6UHN8 A0A0K8S3D6 A0A0K8S4S7 A0A146LVZ5 N6UEG8 A0A0A9Z5I2 A0A1I8PCD8 A0A1I8PC77 A0A0A9X1U0 A0A146KUJ8 A0A154PRE1 B4M8H0 A0A139WDS2 T1H800 A0A182IA55 B4H144 A0A069DY01 A0A1I8MBW9 A0A151WFS6 Q7PS03 A0A182V5U4 A0A195D7R0 A0A182NDR6 A0A151I7K5 A0A151JN89 A0A1I8MBW8 A0A182JGB1 A0A067RJQ3 A0A1I8MBX0 A0A0C9QGJ1 A0A0C9PN45 A0A026WQF9 A0A182SKJ1 A0A3B0KEF7 V9IBI8 A0A224X9R7 V9I9A8
A0A1E1WQJ6 A0A2H1X3B1 A0A385NIS6 A0A2Z5U732 A0A212EN21 A0A0K8V6W5 A0A0K8WJE9 A0A0K8VI49 A0A0K8W2V5 A0A0A1WH39 A0A0A1WIT2 W8AQJ5 W8B0I9 A0A1A9UD14 A0A1B0AH79 A0A1I8M5Y1 A0A1A9Y128 A0A1B0FEI5 A0A1B0C3R6 V5GT10 V5GP54 A0A1B0C696 A0A336MXX7 A0A1A9WHG4 U4UCF2 N6TS49 B4KJH6 A0A232F5K7 D2CG00 A0A139WDV5 A0A182RV52 A0A0A9XLA8 A0A0P8Y4I3 B3MPM8 A0A182LVL1 A0A0A1XIU0 A0A182W4Q6 A0A1B0CML6 A0A182QGM2 B4L379 A0A1S4FMJ9 Q16VL3 A0A034WI75 A0A182Y7H1 A0A0K8UN34 A0A0L0C295 A0A0N0BJK7 A0A182FJQ6 A0A084VJF0 A0A1L8DK46 A0A1L8DXZ7 A0A1L8DJW6 A0A1I8PCF8 A0A1L8DK92 U5EYU3 A0A0K8S3E0 A0A0K8SKT4 N6UHN8 A0A0K8S3D6 A0A0K8S4S7 A0A146LVZ5 N6UEG8 A0A0A9Z5I2 A0A1I8PCD8 A0A1I8PC77 A0A0A9X1U0 A0A146KUJ8 A0A154PRE1 B4M8H0 A0A139WDS2 T1H800 A0A182IA55 B4H144 A0A069DY01 A0A1I8MBW9 A0A151WFS6 Q7PS03 A0A182V5U4 A0A195D7R0 A0A182NDR6 A0A151I7K5 A0A151JN89 A0A1I8MBW8 A0A182JGB1 A0A067RJQ3 A0A1I8MBX0 A0A0C9QGJ1 A0A0C9PN45 A0A026WQF9 A0A182SKJ1 A0A3B0KEF7 V9IBI8 A0A224X9R7 V9I9A8
Pubmed
EMBL
BABH01001863
BABH01001864
BABH01001865
BABH01001866
KQ461196
KPJ06548.1
+ More
KQ459604 KPI92595.1 KZ149999 PZC75350.1 NWSH01000044 PCG80278.1 JTDY01002944 KOB70440.1 GDQN01001810 JAT89244.1 ODYU01013065 SOQ59732.1 MG492013 AYA73999.1 AP017483 BBB06736.1 AGBW02013764 OWR42857.1 GDHF01020819 GDHF01017640 JAI31495.1 JAI34674.1 GDHF01012848 GDHF01001051 JAI39466.1 JAI51263.1 GDHF01013777 JAI38537.1 GDHF01006925 JAI45389.1 GBXI01016090 GBXI01000437 JAC98201.1 JAD13855.1 GBXI01015530 JAC98761.1 GAMC01019808 GAMC01019807 GAMC01019806 JAB86748.1 GAMC01019805 GAMC01019804 GAMC01019803 JAB86751.1 CCAG010014196 JXJN01025092 GALX01005083 JAB63383.1 GALX01005084 JAB63382.1 JXJN01026512 UFQT01001474 SSX30718.1 KB632196 ERL89983.1 APGK01056781 KB741277 ENN71121.1 CH933807 EDW13556.2 NNAY01000911 OXU25955.1 KQ971357 EFA11848.2 KYB26045.1 GBHO01022097 JAG21507.1 CH902620 KPU73871.1 EDV32276.1 KPU73872.1 KPU73873.1 KPU73874.1 KPU73875.1 AXCM01005583 GBXI01003430 JAD10862.1 AJWK01018887 AJWK01018888 AJWK01018889 AJWK01018890 AXCN02001386 CH933810 EDW07007.2 CH477589 EAT38583.1 GAKP01005112 JAC53840.1 GDHF01024313 JAI28001.1 JRES01000984 KNC26440.1 KQ435713 KOX79311.1 ATLV01013553 KE524867 KFB38094.1 GFDF01007339 JAV06745.1 GFDF01002747 JAV11337.1 GFDF01007331 JAV06753.1 GFDF01007330 JAV06754.1 GANO01001810 JAB58061.1 GBRD01018084 JAG47743.1 GBRD01011903 JAG53921.1 APGK01025710 APGK01025711 KB740600 ENN80121.1 GBRD01018089 GBRD01011904 JAG47738.1 GBRD01018087 GBRD01018083 JAG47740.1 GDHC01007404 JAQ11225.1 ENN80120.1 GBHO01030844 GBHO01030842 GBHO01030841 GBHO01006534 GBRD01018090 GBRD01018086 JAG12760.1 JAG12762.1 JAG12763.1 JAG37070.1 JAG47737.1 GBHO01030846 GBHO01030845 GBHO01001040 GBHO01001039 GBRD01018085 GBRD01011902 JAG12758.1 JAG12759.1 JAG42564.1 JAG42565.1 JAG47742.1 GDHC01019802 JAP98826.1 KQ435078 KZC14443.1 CH940654 EDW57496.1 KYB26017.1 ACPB03008313 ACPB03008314 APCN01001124 CH479201 EDW29960.1 GBGD01000124 JAC88765.1 KQ983206 KYQ46679.1 AAAB01008846 EAA06340.6 KQ976760 KYN08494.1 KQ978411 KYM94111.1 KQ978858 KYN27920.1 KK852428 KDR24042.1 GBYB01002584 JAG72351.1 GBYB01002583 JAG72350.1 KK107128 EZA58282.1 OUUW01000011 SPP86700.1 JR037260 AEY57664.1 GFTR01008772 JAW07654.1 JR037261 AEY57665.1
KQ459604 KPI92595.1 KZ149999 PZC75350.1 NWSH01000044 PCG80278.1 JTDY01002944 KOB70440.1 GDQN01001810 JAT89244.1 ODYU01013065 SOQ59732.1 MG492013 AYA73999.1 AP017483 BBB06736.1 AGBW02013764 OWR42857.1 GDHF01020819 GDHF01017640 JAI31495.1 JAI34674.1 GDHF01012848 GDHF01001051 JAI39466.1 JAI51263.1 GDHF01013777 JAI38537.1 GDHF01006925 JAI45389.1 GBXI01016090 GBXI01000437 JAC98201.1 JAD13855.1 GBXI01015530 JAC98761.1 GAMC01019808 GAMC01019807 GAMC01019806 JAB86748.1 GAMC01019805 GAMC01019804 GAMC01019803 JAB86751.1 CCAG010014196 JXJN01025092 GALX01005083 JAB63383.1 GALX01005084 JAB63382.1 JXJN01026512 UFQT01001474 SSX30718.1 KB632196 ERL89983.1 APGK01056781 KB741277 ENN71121.1 CH933807 EDW13556.2 NNAY01000911 OXU25955.1 KQ971357 EFA11848.2 KYB26045.1 GBHO01022097 JAG21507.1 CH902620 KPU73871.1 EDV32276.1 KPU73872.1 KPU73873.1 KPU73874.1 KPU73875.1 AXCM01005583 GBXI01003430 JAD10862.1 AJWK01018887 AJWK01018888 AJWK01018889 AJWK01018890 AXCN02001386 CH933810 EDW07007.2 CH477589 EAT38583.1 GAKP01005112 JAC53840.1 GDHF01024313 JAI28001.1 JRES01000984 KNC26440.1 KQ435713 KOX79311.1 ATLV01013553 KE524867 KFB38094.1 GFDF01007339 JAV06745.1 GFDF01002747 JAV11337.1 GFDF01007331 JAV06753.1 GFDF01007330 JAV06754.1 GANO01001810 JAB58061.1 GBRD01018084 JAG47743.1 GBRD01011903 JAG53921.1 APGK01025710 APGK01025711 KB740600 ENN80121.1 GBRD01018089 GBRD01011904 JAG47738.1 GBRD01018087 GBRD01018083 JAG47740.1 GDHC01007404 JAQ11225.1 ENN80120.1 GBHO01030844 GBHO01030842 GBHO01030841 GBHO01006534 GBRD01018090 GBRD01018086 JAG12760.1 JAG12762.1 JAG12763.1 JAG37070.1 JAG47737.1 GBHO01030846 GBHO01030845 GBHO01001040 GBHO01001039 GBRD01018085 GBRD01011902 JAG12758.1 JAG12759.1 JAG42564.1 JAG42565.1 JAG47742.1 GDHC01019802 JAP98826.1 KQ435078 KZC14443.1 CH940654 EDW57496.1 KYB26017.1 ACPB03008313 ACPB03008314 APCN01001124 CH479201 EDW29960.1 GBGD01000124 JAC88765.1 KQ983206 KYQ46679.1 AAAB01008846 EAA06340.6 KQ976760 KYN08494.1 KQ978411 KYM94111.1 KQ978858 KYN27920.1 KK852428 KDR24042.1 GBYB01002584 JAG72351.1 GBYB01002583 JAG72350.1 KK107128 EZA58282.1 OUUW01000011 SPP86700.1 JR037260 AEY57664.1 GFTR01008772 JAW07654.1 JR037261 AEY57665.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000037510
UP000007151
+ More
UP000078200 UP000092445 UP000095301 UP000092443 UP000092444 UP000092460 UP000091820 UP000030742 UP000019118 UP000009192 UP000215335 UP000007266 UP000075900 UP000007801 UP000075883 UP000075920 UP000092461 UP000075886 UP000008820 UP000076408 UP000037069 UP000053105 UP000069272 UP000030765 UP000095300 UP000076502 UP000008792 UP000015103 UP000075840 UP000008744 UP000075809 UP000007062 UP000075903 UP000078542 UP000075884 UP000078492 UP000075880 UP000027135 UP000053097 UP000075901 UP000268350
UP000078200 UP000092445 UP000095301 UP000092443 UP000092444 UP000092460 UP000091820 UP000030742 UP000019118 UP000009192 UP000215335 UP000007266 UP000075900 UP000007801 UP000075883 UP000075920 UP000092461 UP000075886 UP000008820 UP000076408 UP000037069 UP000053105 UP000069272 UP000030765 UP000095300 UP000076502 UP000008792 UP000015103 UP000075840 UP000008744 UP000075809 UP000007062 UP000075903 UP000078542 UP000075884 UP000078492 UP000075880 UP000027135 UP000053097 UP000075901 UP000268350
PRIDE
ProteinModelPortal
H9JFQ1
A0A194QLW8
A0A194PNA1
A0A2W1BRF3
A0A2A4K8N4
A0A0L7L4U8
+ More
A0A1E1WQJ6 A0A2H1X3B1 A0A385NIS6 A0A2Z5U732 A0A212EN21 A0A0K8V6W5 A0A0K8WJE9 A0A0K8VI49 A0A0K8W2V5 A0A0A1WH39 A0A0A1WIT2 W8AQJ5 W8B0I9 A0A1A9UD14 A0A1B0AH79 A0A1I8M5Y1 A0A1A9Y128 A0A1B0FEI5 A0A1B0C3R6 V5GT10 V5GP54 A0A1B0C696 A0A336MXX7 A0A1A9WHG4 U4UCF2 N6TS49 B4KJH6 A0A232F5K7 D2CG00 A0A139WDV5 A0A182RV52 A0A0A9XLA8 A0A0P8Y4I3 B3MPM8 A0A182LVL1 A0A0A1XIU0 A0A182W4Q6 A0A1B0CML6 A0A182QGM2 B4L379 A0A1S4FMJ9 Q16VL3 A0A034WI75 A0A182Y7H1 A0A0K8UN34 A0A0L0C295 A0A0N0BJK7 A0A182FJQ6 A0A084VJF0 A0A1L8DK46 A0A1L8DXZ7 A0A1L8DJW6 A0A1I8PCF8 A0A1L8DK92 U5EYU3 A0A0K8S3E0 A0A0K8SKT4 N6UHN8 A0A0K8S3D6 A0A0K8S4S7 A0A146LVZ5 N6UEG8 A0A0A9Z5I2 A0A1I8PCD8 A0A1I8PC77 A0A0A9X1U0 A0A146KUJ8 A0A154PRE1 B4M8H0 A0A139WDS2 T1H800 A0A182IA55 B4H144 A0A069DY01 A0A1I8MBW9 A0A151WFS6 Q7PS03 A0A182V5U4 A0A195D7R0 A0A182NDR6 A0A151I7K5 A0A151JN89 A0A1I8MBW8 A0A182JGB1 A0A067RJQ3 A0A1I8MBX0 A0A0C9QGJ1 A0A0C9PN45 A0A026WQF9 A0A182SKJ1 A0A3B0KEF7 V9IBI8 A0A224X9R7 V9I9A8
A0A1E1WQJ6 A0A2H1X3B1 A0A385NIS6 A0A2Z5U732 A0A212EN21 A0A0K8V6W5 A0A0K8WJE9 A0A0K8VI49 A0A0K8W2V5 A0A0A1WH39 A0A0A1WIT2 W8AQJ5 W8B0I9 A0A1A9UD14 A0A1B0AH79 A0A1I8M5Y1 A0A1A9Y128 A0A1B0FEI5 A0A1B0C3R6 V5GT10 V5GP54 A0A1B0C696 A0A336MXX7 A0A1A9WHG4 U4UCF2 N6TS49 B4KJH6 A0A232F5K7 D2CG00 A0A139WDV5 A0A182RV52 A0A0A9XLA8 A0A0P8Y4I3 B3MPM8 A0A182LVL1 A0A0A1XIU0 A0A182W4Q6 A0A1B0CML6 A0A182QGM2 B4L379 A0A1S4FMJ9 Q16VL3 A0A034WI75 A0A182Y7H1 A0A0K8UN34 A0A0L0C295 A0A0N0BJK7 A0A182FJQ6 A0A084VJF0 A0A1L8DK46 A0A1L8DXZ7 A0A1L8DJW6 A0A1I8PCF8 A0A1L8DK92 U5EYU3 A0A0K8S3E0 A0A0K8SKT4 N6UHN8 A0A0K8S3D6 A0A0K8S4S7 A0A146LVZ5 N6UEG8 A0A0A9Z5I2 A0A1I8PCD8 A0A1I8PC77 A0A0A9X1U0 A0A146KUJ8 A0A154PRE1 B4M8H0 A0A139WDS2 T1H800 A0A182IA55 B4H144 A0A069DY01 A0A1I8MBW9 A0A151WFS6 Q7PS03 A0A182V5U4 A0A195D7R0 A0A182NDR6 A0A151I7K5 A0A151JN89 A0A1I8MBW8 A0A182JGB1 A0A067RJQ3 A0A1I8MBX0 A0A0C9QGJ1 A0A0C9PN45 A0A026WQF9 A0A182SKJ1 A0A3B0KEF7 V9IBI8 A0A224X9R7 V9I9A8
PDB
5U74
E-value=2.38185e-150,
Score=1369
Ontologies
KEGG
GO
Topology
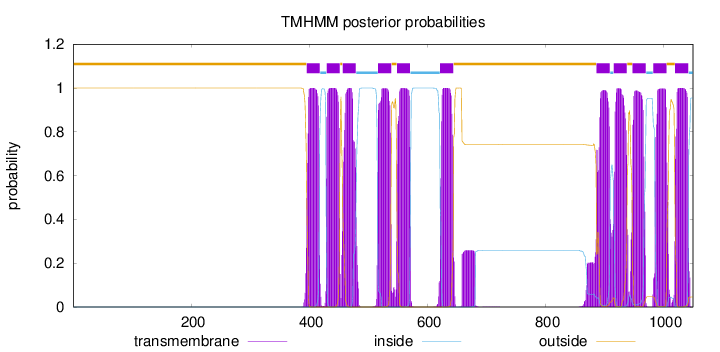
Length:
1050
Number of predicted TMHs:
11
Exp number of AAs in TMHs:
253.89171
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00034
outside
1 - 395
TMhelix
396 - 418
inside
419 - 429
TMhelix
430 - 452
outside
453 - 456
TMhelix
457 - 479
inside
480 - 516
TMhelix
517 - 539
outside
540 - 548
TMhelix
549 - 571
inside
572 - 621
TMhelix
622 - 644
outside
645 - 886
TMhelix
887 - 909
inside
910 - 915
TMhelix
916 - 938
outside
939 - 947
TMhelix
948 - 970
inside
971 - 982
TMhelix
983 - 1005
outside
1006 - 1019
TMhelix
1020 - 1042
inside
1043 - 1050
Population Genetic Test Statistics
Pi
254.698024
Theta
206.234463
Tajima's D
0.738129
CLR
0.023356
CSRT
0.589320533973301
Interpretation
Uncertain
