Gene
KWMTBOMO10894
Pre Gene Modal
BGIBMGA008348
Annotation
PREDICTED:_Niemann-Pick_C1_protein-like_[Papilio_polytes]
Full name
NPC intracellular cholesterol transporter 1 homolog 1b
+ More
NPC intracellular cholesterol transporter 1
NPC intracellular cholesterol transporter 1
Alternative Name
Niemann-Pick type protein homolog 1b
Niemann-Pick C1 protein
Niemann-Pick C1 protein
Location in the cell
PlasmaMembrane Reliability : 4.863
Sequence
CDS
ATGAATTTTTTTATAACGCTCGTTCTTGTGTTTTTGTATACAAGCGATGTGAAGGCGAAATGTCGATTTCGCGGTGAGTGCATTGAAATAGCAGGCCATGCAAAACCTTGCCCTGTGGATATTGAAGCAAGACCTGTCGTAGAAGGACTGTCGCCAGCAGATGCCGAAGAAATCGTTGACATTTTAACTGAAAGATGTCCTAGTCTTGTTTTCGATGAAAATGGTGATAAGAAGCCATTCAATGAAATCTTAACTTGCTGTGAACCCGTCCAAATCAGGAAGCTATCGGAGAGCTTAATGTTGGCCGATGGTATCCTGGGCCGTTGTCCAACATGCTTAAGAAATTTCGCCAGACAAATTTGTGAGATGAACTGTTCTCCTGATCAATCTCGTTTCGTTGATGTCTACACTGAAGTATCAGGAGGTGTCCGATATGTGAACGAAATCGATTATAGACTTTATGAACCGTTTATGTTGGGAGCTCACGCATCGTGTGCCGGCGTTATAATTCCACAAACGGGCTTACCAGCAATTAACATGATGTGTGGCAATGCAGTTGTTTGTGATGCTGATGCATGGTTTGGATTTACTGGAGATACCTCAAGCAATCCAATGGCACCGGTGCAAGTTAACTTTTTAAGGTGGCCTACACCCGAAGACTCAATGAATGCGCCTGCTCCACAGTGTAACGAAACCGCAGAAGGTGATCTTCCATGTAGTTGCGTTGATTGCGCTGCGATGTGCCCAACTGGGACTGAGCCCGTTGTTCCTGAAATTTGTACTGTTCTAGACGTTCACTGCGTTGGTTTCGCAGTTGGTATTACGGTATTTGTAATAACAACTATAATATTCATTATTTTAACGCTACTAGAATACCGTAAGCTAAGGAAAGTTGAAACCAAGACTCCGTTACCGGCCGATGTTCAAAGGTTTACAGTATTTTTCCAAAATCTATTTGCGAAGATTGGTGTTTTTGCCGCAAACAATCCTATACTTATAATAATGGTAACCTCGTGGGTATCAATAGGTGTGTTATATGGCGCTTTTAGTCTTAACATAACTTCGAATCCTATTGAACTTTGGTCTTCACCAGTATCCCGCAGTCGAGAGCAATTAAATTATTTCAATACTAGATTTGGTCCTTTTTACCGTGCGGCACAAGTTTTCTTGCAGTTCAAAGGATTGGAGCCATTTGAAGTAGATAATGTGACATATGGATCTGCATTTAGAATTGAAGCTATTCATGAATTGATCAAATTAGAAGATGAAATTTATAAAATTGGAAAAGATGACGGTGGTGTTTCACTAGAACAGGTTTGTTATGCTCCTAATCGTCCCAGAGGGGGAGAACAAAGACTTGATCAATGTGTTTCGATGTCTGTATCTGTATACTTCGGTCAGGATAGGGATAACATCAATGAAAATACCTATTTAGATCAAATACAAAATTGTCTGAACAATCACTATGCACTCAACTGCTTAGCATCATGGGGAGGTGGTTCCGAACCTGAGATTACTTTTGGAGGTTTCGAAGGAAACAACATTCTTAGTGCTGACACGCTTCTTATTAACTTTCCAATTACAAACTTCATGTTAGAGGAAGATTTGAGACCTGTCTTGGAATGGGAGCAGAAATTCATAGATCTATTGCATGACTACAGAGACAACTGGAAGTCAGATTTCGTTGAAGTGGCTTTCGGAACCGAAAGATCAATCGAAGATGAGATTCGACGGGTTTCAGTTGCGGAAATTTTGCCGATTTCGATAAGCTACATTATAATGTTTATATATGTTATTATAGCGTTAGGAAATGTAAGGCACTGTAAGACGTGGTTACTTGATAGTAAAATAGTAGTAGCAGCTGGTAGTATTGTCGTAGTACTCCTTTCCATTGGATGTGCTTTAGGCTTAATGGGCTACACAGGTATTACAACAACCTTATTGGCCATTAATGTTATACCTTTCTTCATTCTTTCAGTTGGTATTGACAACGTTTTCTTAATGGTGAATACATTGCACGATATTCAGAACGATCTTAAATCATACGATGACTATAACGAAAATTTTAGTTTTTCAAAGAAACGTAGTTTCGTTTTTGAAAAGATGATGAGTGCAGTCGGCCCTTCCATATTTGTTTCATCAGTAACCCAAATTACTTGTTTTGGTATTGGAAGTATAACTAATTTCCCAGCTGTTCAAACATTTGCTGTATTTGCGAGTTTTTCCTTAACTTTCCTATTTGTATTTCAAATCACGACTGTGGTAGCGATTTTGTCCCTTGATTACAAACGTGTGTCGCAAAACCGCCCTGATCTATTTTGCTGTATTCGAAAGAAGATTCTTAATGATGACGATCCTTTAAATTCTGAAACTCCTTATAAGAGTATCACGAAAAGACTCATGAAGCCTTATTCTAAATTTTTACTAAACTGGAAGACAAAGATCATTGTTGCTATTTTGTTTATGCTGTTGGTATCTACTAGTGTTGTTTTGATTCCACAAGTGGAAATTGGTTTAGACCAAGAAATGAGTTTGCCAACCGATTCTTATGTATATAGTTATTTAGTAGCAGTTAGTAAGCTTTTACGTCTTGGACCCCCGGTATATTTTGTTCTTAAGGCTGGCTTAGACTTTTCAAACACAAATCATCAAAATGTTATCTGCGGTGGTCAATTATGTTATGATGATTCTCTATTCACTCAAGTGTTTTTAGCTTCTCAACATAGCAGCATCACTAGGATTTCAAAAAGTTCTAATTCATGTACAGATAACTCCCCTGAATGTGCTTTCTGTACTATAGGAAGGGACGAATGGGCCAACGGATTACGTCCCAGTTCAGAAGCGTTTAAGAGATATATTCCATTCTTTTTGCGAGATGAACCGACTCCAACCTGCAATAAAGGGGGTCTTGCGAGTTATGCAAATGCCGTGAATTATTTACTTGATTCTGAAGGACATGCCACTGTGCGTGACACTAATTTCATGGCTTACCACACTTCAGTAAGTACATCTTCAGATTATATCGAAGCAGTAAAGTATGGATATGAAATCAGTGATAGTATAACTGCAGCCATAAAGAAACGCACAGGTCTAGATGTGGAAGTGTTTCCATATTCAATATTTTACGTTTACTATGAACAATACCTAACCATATGGGGTGATACGTTTGCAGCTCTAGGCTATTGTCTCATTGGTGCATTGGTTATCAATTTACTAGCATCTGGATTCAATTTCTTAACAACTTTTGCCGTTATATTTACGGCGATTTTGGTTGTCATAAACATGATGGGCGTTATGTACATTTGGAATATTCCTTTAAACGCTGTGTCTTGTGTTAACTTAGTTGTTTCTATCGGTATTGCTGTTGAGTTCTGCAGTCACATTGCCTATGCATTCGATTCGAGTACTCGTCCCGCAGATGAAAGAGTACAAGATGCGTTACAAAAGATAGGTTCGACAGTTATTACTGGTATTACATTTACGAATATACCTATTATTGTTTTAGCGTTCTCATACACGCAAGTAATAGAGGTATTTTTCTTTAGAATGTTCTTTACTTTGATAATATTGGGATTCTTACACGGGATGGTCTTCTTCCCTGTATTGTTAAGCTACCTGAACAATTTAAGGAACAAATAA
Protein
MNFFITLVLVFLYTSDVKAKCRFRGECIEIAGHAKPCPVDIEARPVVEGLSPADAEEIVDILTERCPSLVFDENGDKKPFNEILTCCEPVQIRKLSESLMLADGILGRCPTCLRNFARQICEMNCSPDQSRFVDVYTEVSGGVRYVNEIDYRLYEPFMLGAHASCAGVIIPQTGLPAINMMCGNAVVCDADAWFGFTGDTSSNPMAPVQVNFLRWPTPEDSMNAPAPQCNETAEGDLPCSCVDCAAMCPTGTEPVVPEICTVLDVHCVGFAVGITVFVITTIIFIILTLLEYRKLRKVETKTPLPADVQRFTVFFQNLFAKIGVFAANNPILIIMVTSWVSIGVLYGAFSLNITSNPIELWSSPVSRSREQLNYFNTRFGPFYRAAQVFLQFKGLEPFEVDNVTYGSAFRIEAIHELIKLEDEIYKIGKDDGGVSLEQVCYAPNRPRGGEQRLDQCVSMSVSVYFGQDRDNINENTYLDQIQNCLNNHYALNCLASWGGGSEPEITFGGFEGNNILSADTLLINFPITNFMLEEDLRPVLEWEQKFIDLLHDYRDNWKSDFVEVAFGTERSIEDEIRRVSVAEILPISISYIIMFIYVIIALGNVRHCKTWLLDSKIVVAAGSIVVVLLSIGCALGLMGYTGITTTLLAINVIPFFILSVGIDNVFLMVNTLHDIQNDLKSYDDYNENFSFSKKRSFVFEKMMSAVGPSIFVSSVTQITCFGIGSITNFPAVQTFAVFASFSLTFLFVFQITTVVAILSLDYKRVSQNRPDLFCCIRKKILNDDDPLNSETPYKSITKRLMKPYSKFLLNWKTKIIVAILFMLLVSTSVVLIPQVEIGLDQEMSLPTDSYVYSYLVAVSKLLRLGPPVYFVLKAGLDFSNTNHQNVICGGQLCYDDSLFTQVFLASQHSSITRISKSSNSCTDNSPECAFCTIGRDEWANGLRPSSEAFKRYIPFFLRDEPTPTCNKGGLASYANAVNYLLDSEGHATVRDTNFMAYHTSVSTSSDYIEAVKYGYEISDSITAAIKKRTGLDVEVFPYSIFYVYYEQYLTIWGDTFAALGYCLIGALVINLLASGFNFLTTFAVIFTAILVVINMMGVMYIWNIPLNAVSCVNLVVSIGIAVEFCSHIAYAFDSSTRPADERVQDALQKIGSTVITGITFTNIPIIVLAFSYTQVIEVFFFRMFFTLIILGFLHGMVFFPVLLSYLNNLRNK
Summary
Description
Important for cholesterol absorption at the midgut epithelium. Acts only in the early steps of sterol absorption, prior to Npc1a-dependent intracellular sterol trafficking.
Intracellular cholesterol transporter which acts in concert with NPC2 and plays an important role in the egress of cholesterol from the endosomal/lysosomal compartment (PubMed:9211849, PubMed:9927649, PubMed:10821832, PubMed:18772377, PubMed:27238017, PubMed:12554680). Unesterified cholesterol that has been released from LDLs in the lumen of the late endosomes/lysosomes is transferred by NPC2 to the cholesterol-binding pocket in the N-terminal domain of NPC1 (PubMed:9211849, PubMed:9927649, PubMed:18772377, PubMed:19563754, PubMed:27238017, PubMed:28784760). Cholesterol binds to NPC1 with the hydroxyl group buried in the binding pocket (PubMed:19563754). Binds oxysterol with higher affinity than cholesterol. May play a role in vesicular trafficking in glia, a process that may be crucial for maintaining the structural and functional integrity of nerve terminals (Probable).
(Microbial infection) Acts as an endosomal entry receptor for ebolavirus.
Intracellular cholesterol transporter which acts in concert with NPC2 and plays an important role in the egress of cholesterol from the endosomal/lysosomal compartment (PubMed:9211849, PubMed:9927649, PubMed:10821832, PubMed:18772377, PubMed:27238017, PubMed:12554680). Unesterified cholesterol that has been released from LDLs in the lumen of the late endosomes/lysosomes is transferred by NPC2 to the cholesterol-binding pocket in the N-terminal domain of NPC1 (PubMed:9211849, PubMed:9927649, PubMed:18772377, PubMed:19563754, PubMed:27238017, PubMed:28784760). Cholesterol binds to NPC1 with the hydroxyl group buried in the binding pocket (PubMed:19563754). Binds oxysterol with higher affinity than cholesterol. May play a role in vesicular trafficking in glia, a process that may be crucial for maintaining the structural and functional integrity of nerve terminals (Probable).
(Microbial infection) Acts as an endosomal entry receptor for ebolavirus.
Subunit
Interacts (via the second lumenal domain) with NPC2 (PubMed:18772377, PubMed:27238017, PubMed:27551080). Interacts with TMEM97 (PubMed:19583955). Interacts with TIM1 (PubMed:25855742).
(Microbial infection) Interacts with ebolavirus glycoprotein.
(Microbial infection) Interacts with ebolavirus glycoprotein.
Similarity
Belongs to the patched family.
Keywords
Cell membrane
Cholesterol metabolism
Complete proteome
Disulfide bond
Glycoprotein
Lipid metabolism
Membrane
Reference proteome
Signal
Steroid metabolism
Sterol metabolism
Transmembrane
Transmembrane helix
3D-structure
Alternative splicing
Disease mutation
Endosome
Host cell receptor for virus entry
Host-virus interaction
Lysosome
Niemann-Pick disease
Polymorphism
Receptor
Feature
chain NPC intracellular cholesterol transporter 1 homolog 1b
splice variant In isoform 2.
sequence variant In NPC1; dbSNP:rs747049347.
splice variant In isoform 2.
sequence variant In NPC1; dbSNP:rs747049347.
Uniprot
A0A194QLW8
A0A194PNA1
A0A2A4K8N4
A0A2W1BRF3
A0A0L7L4U8
H9JFQ1
+ More
A0A1E1WQJ6 A0A2H1X3B1 A0A2Z5U732 A0A385NIS6 A0A212EN21 A0A336MXX7 A0A182RV52 A0A182W4Q6 A0A182LVL1 A0A182XLP3 A0A182SKJ1 Q16VL3 A0A182NDR6 A0A182FJQ6 A0A0L0C295 A0A182U4J9 A0A182PQ62 A0A084VJF0 A0A1I8M5Y1 B4L379 A0A182JGB1 A0A1I8PTT6 A0A1L8DY99 A0A1L8DYC2 A0A1W4VJ55 A0A1L8DYR8 A0A182K6I0 Q9VRC9 A0A151ITF2 W8BF01 A0A139WDV5 A0A151WFS6 A0A195EWZ0 A0A195D7R0 A0A1Y1JY67 F4WWI9 A0A232F5K7 A0A1Y1JRU7 A0A1A9WHG4 V9IBI8 A0A2A3EN71 V9I9A8 N6UEG8 E0W1Y7 E0VVZ7 A0A084W877 N6UHN8 A0A182H2F2 A0A3Q0FVZ6 A0A151PHK5 N6TS49 A0A0N5DXQ8 A0A1B0CML6 A0A1V4KGB4 A0A1V4KG21 A0A099YPK4 A0A1V4KG51 A0A091EX33 T1E5N3 A0A250Y7G1 A0A3Q2IC60 F4WKN5 A0A1L8DK92 A0A1L8DJW6 A0A1L8DK46 A0A1L8DXZ7 M3Y344 U6CRS1 A0A091PJJ9 A0A2Y9IR23 A0A1S3WHY8 F1NQT4 U3CBB5 F7INK6 M3VUX6 A0A2I2UCH5 U3FMJ3 A0A0A8JCD0 F7INM5 A0A2K5CV08 A0A2K6UKX3 A0A193DRS0 O15118 F1PB50 Q8MI49 A0A2K6UL71 Q59GR1 Q8MKD8 A0A3Q2HH91 A0A2K5CV07 A0A3Q7TM00 J9K7Y2 W5M0D1 A0A1S3F160
A0A1E1WQJ6 A0A2H1X3B1 A0A2Z5U732 A0A385NIS6 A0A212EN21 A0A336MXX7 A0A182RV52 A0A182W4Q6 A0A182LVL1 A0A182XLP3 A0A182SKJ1 Q16VL3 A0A182NDR6 A0A182FJQ6 A0A0L0C295 A0A182U4J9 A0A182PQ62 A0A084VJF0 A0A1I8M5Y1 B4L379 A0A182JGB1 A0A1I8PTT6 A0A1L8DY99 A0A1L8DYC2 A0A1W4VJ55 A0A1L8DYR8 A0A182K6I0 Q9VRC9 A0A151ITF2 W8BF01 A0A139WDV5 A0A151WFS6 A0A195EWZ0 A0A195D7R0 A0A1Y1JY67 F4WWI9 A0A232F5K7 A0A1Y1JRU7 A0A1A9WHG4 V9IBI8 A0A2A3EN71 V9I9A8 N6UEG8 E0W1Y7 E0VVZ7 A0A084W877 N6UHN8 A0A182H2F2 A0A3Q0FVZ6 A0A151PHK5 N6TS49 A0A0N5DXQ8 A0A1B0CML6 A0A1V4KGB4 A0A1V4KG21 A0A099YPK4 A0A1V4KG51 A0A091EX33 T1E5N3 A0A250Y7G1 A0A3Q2IC60 F4WKN5 A0A1L8DK92 A0A1L8DJW6 A0A1L8DK46 A0A1L8DXZ7 M3Y344 U6CRS1 A0A091PJJ9 A0A2Y9IR23 A0A1S3WHY8 F1NQT4 U3CBB5 F7INK6 M3VUX6 A0A2I2UCH5 U3FMJ3 A0A0A8JCD0 F7INM5 A0A2K5CV08 A0A2K6UKX3 A0A193DRS0 O15118 F1PB50 Q8MI49 A0A2K6UL71 Q59GR1 Q8MKD8 A0A3Q2HH91 A0A2K5CV07 A0A3Q7TM00 J9K7Y2 W5M0D1 A0A1S3F160
Pubmed
26354079
28756777
26227816
19121390
22118469
17510324
+ More
26108605 24438588 25315136 17994087 10731132 12537572 17339027 24495485 18362917 19820115 28004739 21719571 28648823 23537049 20566863 26483478 22293439 23758969 25727380 28087693 19892987 15592404 25243066 17975172 9211849 10425213 11754101 14702039 16177791 15489334 9927649 10821832 17897319 18772377 19583955 19159218 18832164 20674853 21269460 21412152 21866103 25855742 25944712 19563754 26771495 27238017 26846330 27551080 28784760 9634529 10521290 10521297 10480349 11182931 11349231 11333381 11545687 11479732 12408188 12401890 12554680 12955717 16098014 15774455 16126423 16802107 23453666 16341006 12809639
26108605 24438588 25315136 17994087 10731132 12537572 17339027 24495485 18362917 19820115 28004739 21719571 28648823 23537049 20566863 26483478 22293439 23758969 25727380 28087693 19892987 15592404 25243066 17975172 9211849 10425213 11754101 14702039 16177791 15489334 9927649 10821832 17897319 18772377 19583955 19159218 18832164 20674853 21269460 21412152 21866103 25855742 25944712 19563754 26771495 27238017 26846330 27551080 28784760 9634529 10521290 10521297 10480349 11182931 11349231 11333381 11545687 11479732 12408188 12401890 12554680 12955717 16098014 15774455 16126423 16802107 23453666 16341006 12809639
EMBL
KQ461196
KPJ06548.1
KQ459604
KPI92595.1
NWSH01000044
PCG80278.1
+ More
KZ149999 PZC75350.1 JTDY01002944 KOB70440.1 BABH01001863 BABH01001864 BABH01001865 BABH01001866 GDQN01001810 JAT89244.1 ODYU01013065 SOQ59732.1 AP017483 BBB06736.1 MG492013 AYA73999.1 AGBW02013764 OWR42857.1 UFQT01001474 SSX30718.1 AXCM01005583 CH477589 EAT38583.1 JRES01000984 KNC26440.1 ATLV01013553 KE524867 KFB38094.1 CH933810 EDW07007.2 GFDF01002661 JAV11423.1 GFDF01002660 JAV11424.1 GFDF01002669 JAV11415.1 AE014298 KQ981019 KYN10247.1 GAMC01006635 JAB99920.1 KQ971357 KYB26045.1 KQ983206 KYQ46679.1 KQ981953 KYN32399.1 KQ976760 KYN08494.1 GEZM01102128 JAV51946.1 GL888408 EGI61420.1 NNAY01000911 OXU25955.1 GEZM01102125 JAV51949.1 JR037260 AEY57664.1 KZ288215 PBC32491.1 JR037261 AEY57665.1 APGK01025710 APGK01025711 KB740600 ENN80120.1 DS235873 EEB19581.1 DS235816 EEB17553.1 ATLV01021379 ATLV01021380 ATLV01021381 ATLV01021382 KE525317 KFB46421.1 ENN80121.1 JXUM01024284 KQ560685 KXJ81281.1 AKHW03000230 KYO48265.1 APGK01056781 KB741277 ENN71121.1 AJWK01018887 AJWK01018888 AJWK01018889 AJWK01018890 LSYS01003169 OPJ83373.1 OPJ83374.1 KL884815 KGL72244.1 OPJ83375.1 KK719137 KFO61541.1 GAAZ01001621 GBKC01002383 GBKD01001446 JAA96322.1 JAG43687.1 JAG46172.1 GFFV01000432 JAV39513.1 GL888206 EGI65127.1 GFDF01007330 JAV06754.1 GFDF01007331 JAV06753.1 GFDF01007339 JAV06745.1 GFDF01002747 JAV11337.1 AEYP01011843 HAAF01001168 CCP72994.1 KL391117 KFP91660.1 AADN05000027 GAMT01000689 GAMT01000688 GAMT01000687 GAMS01000344 GAMS01000343 GAMS01000342 JAB11172.1 JAB22793.1 AANG04003548 GAMR01006953 GAMR01006952 GAMR01006951 GAMQ01004369 GAMQ01004368 GAMQ01004367 GAMP01006860 GAMP01006859 JAB26979.1 JAB37482.1 JAB45896.1 AB971140 BAQ08368.1 KU695564 ANN44507.1 AF002020 AF157379 AF157365 AF157366 AF157367 AF157368 AF157369 AF157370 AF157371 AF157372 AF157373 AF157374 AF157375 AF157376 AF157377 AF157378 AF338230 AH009108 AK293779 AC010853 AC026634 BC063302 AAEX03005455 AF503633 AAM27450.1 AB209048 BAD92285.1 AF503634 AAM27451.1 ABLF02019318 ABLF02019320 ABLF02019324 AHAT01004140
KZ149999 PZC75350.1 JTDY01002944 KOB70440.1 BABH01001863 BABH01001864 BABH01001865 BABH01001866 GDQN01001810 JAT89244.1 ODYU01013065 SOQ59732.1 AP017483 BBB06736.1 MG492013 AYA73999.1 AGBW02013764 OWR42857.1 UFQT01001474 SSX30718.1 AXCM01005583 CH477589 EAT38583.1 JRES01000984 KNC26440.1 ATLV01013553 KE524867 KFB38094.1 CH933810 EDW07007.2 GFDF01002661 JAV11423.1 GFDF01002660 JAV11424.1 GFDF01002669 JAV11415.1 AE014298 KQ981019 KYN10247.1 GAMC01006635 JAB99920.1 KQ971357 KYB26045.1 KQ983206 KYQ46679.1 KQ981953 KYN32399.1 KQ976760 KYN08494.1 GEZM01102128 JAV51946.1 GL888408 EGI61420.1 NNAY01000911 OXU25955.1 GEZM01102125 JAV51949.1 JR037260 AEY57664.1 KZ288215 PBC32491.1 JR037261 AEY57665.1 APGK01025710 APGK01025711 KB740600 ENN80120.1 DS235873 EEB19581.1 DS235816 EEB17553.1 ATLV01021379 ATLV01021380 ATLV01021381 ATLV01021382 KE525317 KFB46421.1 ENN80121.1 JXUM01024284 KQ560685 KXJ81281.1 AKHW03000230 KYO48265.1 APGK01056781 KB741277 ENN71121.1 AJWK01018887 AJWK01018888 AJWK01018889 AJWK01018890 LSYS01003169 OPJ83373.1 OPJ83374.1 KL884815 KGL72244.1 OPJ83375.1 KK719137 KFO61541.1 GAAZ01001621 GBKC01002383 GBKD01001446 JAA96322.1 JAG43687.1 JAG46172.1 GFFV01000432 JAV39513.1 GL888206 EGI65127.1 GFDF01007330 JAV06754.1 GFDF01007331 JAV06753.1 GFDF01007339 JAV06745.1 GFDF01002747 JAV11337.1 AEYP01011843 HAAF01001168 CCP72994.1 KL391117 KFP91660.1 AADN05000027 GAMT01000689 GAMT01000688 GAMT01000687 GAMS01000344 GAMS01000343 GAMS01000342 JAB11172.1 JAB22793.1 AANG04003548 GAMR01006953 GAMR01006952 GAMR01006951 GAMQ01004369 GAMQ01004368 GAMQ01004367 GAMP01006860 GAMP01006859 JAB26979.1 JAB37482.1 JAB45896.1 AB971140 BAQ08368.1 KU695564 ANN44507.1 AF002020 AF157379 AF157365 AF157366 AF157367 AF157368 AF157369 AF157370 AF157371 AF157372 AF157373 AF157374 AF157375 AF157376 AF157377 AF157378 AF338230 AH009108 AK293779 AC010853 AC026634 BC063302 AAEX03005455 AF503633 AAM27450.1 AB209048 BAD92285.1 AF503634 AAM27451.1 ABLF02019318 ABLF02019320 ABLF02019324 AHAT01004140
Proteomes
UP000053240
UP000053268
UP000218220
UP000037510
UP000005204
UP000007151
+ More
UP000075900 UP000075920 UP000075883 UP000076407 UP000075901 UP000008820 UP000075884 UP000069272 UP000037069 UP000075902 UP000075885 UP000030765 UP000095301 UP000009192 UP000075880 UP000095300 UP000192221 UP000075881 UP000000803 UP000078492 UP000007266 UP000075809 UP000078541 UP000078542 UP000007755 UP000215335 UP000091820 UP000242457 UP000019118 UP000009046 UP000069940 UP000249989 UP000189705 UP000050525 UP000046395 UP000092461 UP000190648 UP000053641 UP000052976 UP000002281 UP000000715 UP000248482 UP000079721 UP000000539 UP000008225 UP000011712 UP000233020 UP000233220 UP000005640 UP000002254 UP000286640 UP000007819 UP000018468 UP000081671
UP000075900 UP000075920 UP000075883 UP000076407 UP000075901 UP000008820 UP000075884 UP000069272 UP000037069 UP000075902 UP000075885 UP000030765 UP000095301 UP000009192 UP000075880 UP000095300 UP000192221 UP000075881 UP000000803 UP000078492 UP000007266 UP000075809 UP000078541 UP000078542 UP000007755 UP000215335 UP000091820 UP000242457 UP000019118 UP000009046 UP000069940 UP000249989 UP000189705 UP000050525 UP000046395 UP000092461 UP000190648 UP000053641 UP000052976 UP000002281 UP000000715 UP000248482 UP000079721 UP000000539 UP000008225 UP000011712 UP000233020 UP000233220 UP000005640 UP000002254 UP000286640 UP000007819 UP000018468 UP000081671
ProteinModelPortal
A0A194QLW8
A0A194PNA1
A0A2A4K8N4
A0A2W1BRF3
A0A0L7L4U8
H9JFQ1
+ More
A0A1E1WQJ6 A0A2H1X3B1 A0A2Z5U732 A0A385NIS6 A0A212EN21 A0A336MXX7 A0A182RV52 A0A182W4Q6 A0A182LVL1 A0A182XLP3 A0A182SKJ1 Q16VL3 A0A182NDR6 A0A182FJQ6 A0A0L0C295 A0A182U4J9 A0A182PQ62 A0A084VJF0 A0A1I8M5Y1 B4L379 A0A182JGB1 A0A1I8PTT6 A0A1L8DY99 A0A1L8DYC2 A0A1W4VJ55 A0A1L8DYR8 A0A182K6I0 Q9VRC9 A0A151ITF2 W8BF01 A0A139WDV5 A0A151WFS6 A0A195EWZ0 A0A195D7R0 A0A1Y1JY67 F4WWI9 A0A232F5K7 A0A1Y1JRU7 A0A1A9WHG4 V9IBI8 A0A2A3EN71 V9I9A8 N6UEG8 E0W1Y7 E0VVZ7 A0A084W877 N6UHN8 A0A182H2F2 A0A3Q0FVZ6 A0A151PHK5 N6TS49 A0A0N5DXQ8 A0A1B0CML6 A0A1V4KGB4 A0A1V4KG21 A0A099YPK4 A0A1V4KG51 A0A091EX33 T1E5N3 A0A250Y7G1 A0A3Q2IC60 F4WKN5 A0A1L8DK92 A0A1L8DJW6 A0A1L8DK46 A0A1L8DXZ7 M3Y344 U6CRS1 A0A091PJJ9 A0A2Y9IR23 A0A1S3WHY8 F1NQT4 U3CBB5 F7INK6 M3VUX6 A0A2I2UCH5 U3FMJ3 A0A0A8JCD0 F7INM5 A0A2K5CV08 A0A2K6UKX3 A0A193DRS0 O15118 F1PB50 Q8MI49 A0A2K6UL71 Q59GR1 Q8MKD8 A0A3Q2HH91 A0A2K5CV07 A0A3Q7TM00 J9K7Y2 W5M0D1 A0A1S3F160
A0A1E1WQJ6 A0A2H1X3B1 A0A2Z5U732 A0A385NIS6 A0A212EN21 A0A336MXX7 A0A182RV52 A0A182W4Q6 A0A182LVL1 A0A182XLP3 A0A182SKJ1 Q16VL3 A0A182NDR6 A0A182FJQ6 A0A0L0C295 A0A182U4J9 A0A182PQ62 A0A084VJF0 A0A1I8M5Y1 B4L379 A0A182JGB1 A0A1I8PTT6 A0A1L8DY99 A0A1L8DYC2 A0A1W4VJ55 A0A1L8DYR8 A0A182K6I0 Q9VRC9 A0A151ITF2 W8BF01 A0A139WDV5 A0A151WFS6 A0A195EWZ0 A0A195D7R0 A0A1Y1JY67 F4WWI9 A0A232F5K7 A0A1Y1JRU7 A0A1A9WHG4 V9IBI8 A0A2A3EN71 V9I9A8 N6UEG8 E0W1Y7 E0VVZ7 A0A084W877 N6UHN8 A0A182H2F2 A0A3Q0FVZ6 A0A151PHK5 N6TS49 A0A0N5DXQ8 A0A1B0CML6 A0A1V4KGB4 A0A1V4KG21 A0A099YPK4 A0A1V4KG51 A0A091EX33 T1E5N3 A0A250Y7G1 A0A3Q2IC60 F4WKN5 A0A1L8DK92 A0A1L8DJW6 A0A1L8DK46 A0A1L8DXZ7 M3Y344 U6CRS1 A0A091PJJ9 A0A2Y9IR23 A0A1S3WHY8 F1NQT4 U3CBB5 F7INK6 M3VUX6 A0A2I2UCH5 U3FMJ3 A0A0A8JCD0 F7INM5 A0A2K5CV08 A0A2K6UKX3 A0A193DRS0 O15118 F1PB50 Q8MI49 A0A2K6UL71 Q59GR1 Q8MKD8 A0A3Q2HH91 A0A2K5CV07 A0A3Q7TM00 J9K7Y2 W5M0D1 A0A1S3F160
PDB
5U74
E-value=4.0088e-173,
Score=1566
Ontologies
KEGG
GO
GO:0016021
GO:0005319
GO:0007391
GO:0007417
GO:0030299
GO:0007422
GO:0005886
GO:0008203
GO:0007628
GO:0031579
GO:0015485
GO:0005635
GO:0016242
GO:0005768
GO:0005794
GO:0033344
GO:0071383
GO:0045121
GO:0042632
GO:1905103
GO:0006897
GO:0005576
GO:0046718
GO:0032367
GO:0006914
GO:0005887
GO:0006486
GO:0090150
GO:0005783
GO:0071404
GO:0042493
GO:0048471
GO:0005764
GO:0004888
GO:0034383
GO:0007041
GO:0015248
GO:0001618
GO:0008206
GO:0060548
GO:0046686
GO:0005765
GO:0030301
GO:0070062
GO:0016020
GO:0031902
GO:0038023
GO:0035855
GO:0055113
GO:0043009
GO:0001666
GO:0006508
GO:0043565
GO:0003676
GO:0005515
GO:0051028
GO:0005622
GO:0006810
GO:0003824
Topology
Subcellular location
Cell membrane
Late endosome membrane
Lysosome membrane
Late endosome membrane
Lysosome membrane
SignalP
Position: 1 - 19,
Likelihood: 0.994720
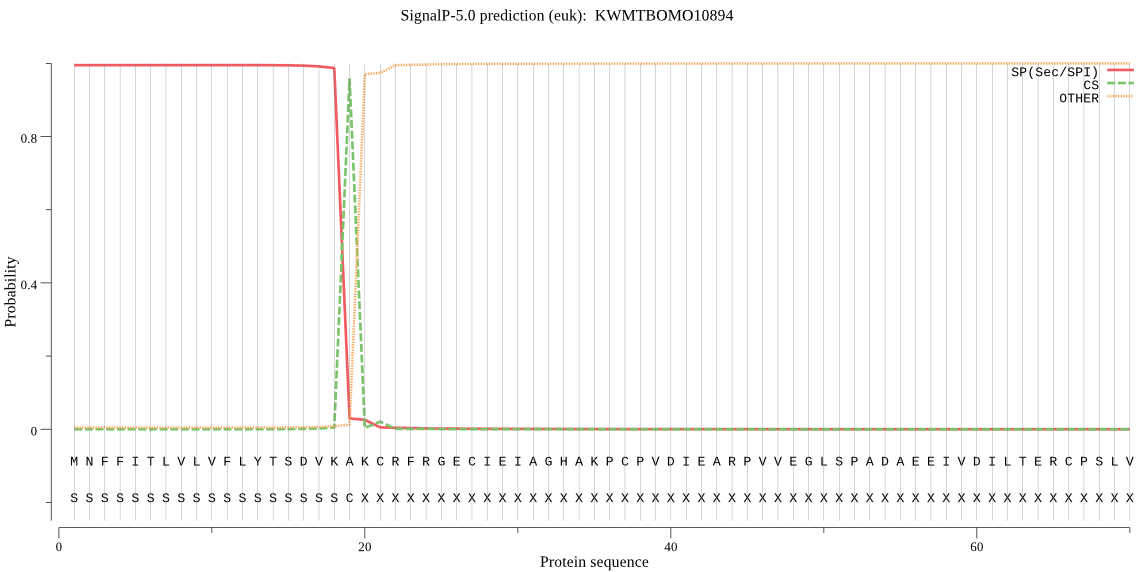
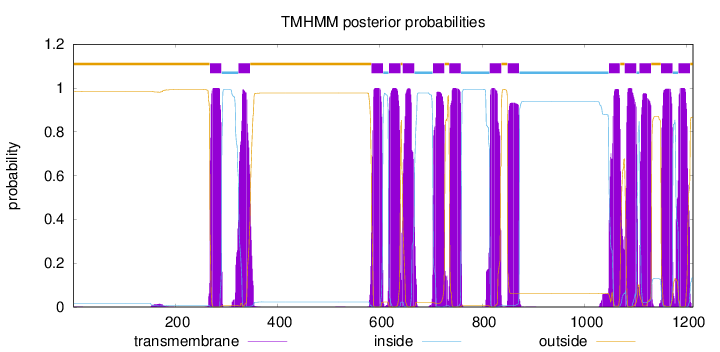
Length:
1212
Number of predicted TMHs:
14
Exp number of AAs in TMHs:
312.66185
Exp number, first 60 AAs:
0.00804
Total prob of N-in:
0.01626
outside
1 - 267
TMhelix
268 - 290
inside
291 - 323
TMhelix
324 - 346
outside
347 - 583
TMhelix
584 - 606
inside
607 - 617
TMhelix
618 - 640
outside
641 - 644
TMhelix
645 - 667
inside
668 - 703
TMhelix
704 - 726
outside
727 - 735
TMhelix
736 - 758
inside
759 - 814
TMhelix
815 - 837
outside
838 - 849
TMhelix
850 - 872
inside
873 - 1047
TMhelix
1048 - 1069
outside
1070 - 1078
TMhelix
1079 - 1101
inside
1102 - 1107
TMhelix
1108 - 1130
outside
1131 - 1149
TMhelix
1150 - 1172
inside
1173 - 1183
TMhelix
1184 - 1206
outside
1207 - 1212
Population Genetic Test Statistics
Pi
249.90183
Theta
224.601551
Tajima's D
0.295551
CLR
0.245024
CSRT
0.451427428628569
Interpretation
Uncertain
