Pre Gene Modal
BGIBMGA008350
Annotation
PREDICTED:_uncharacterized_protein_LOC101744659_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 3.685
Sequence
CDS
ATGGATTTCAGGATTAAAATATATCTATTTATGTCTTGTTTGTATTGTTCAATATTTCCAATTTACGGTGAAGATGTGTGCCTGAAAAAAAGTGCGTGCGTTTGTGAGTTTTCCAACGGCACCGGAATAGATTTGTCTTCGGCCAGTAAATCGACATTTTTTTCTGCTCAAACTTTTGAAGTAGCTCCAGCAACAAATGCGATTGAACTTAGTACTTATTACTATCATCCTTGCTATGACATACTTCTAAAACCGAATTCTACAACTGTTAACAATACTTGTACAGTACCGACAGCCATTTGCAGACACAGAGAAGTGCTTCATGCTGTTGATGGTACACCAGACACATATAAAAAGGATCTAAATGTGTATGATTTAATTGGCAATACTGATAAATCGGTATTTAGTGCTAAAGGAGATGCAATAGTTTACACTCATGGTCCATCAAATACAACAGTTCTCCTCATTTGCTCTCTGTCTGATGGTGAATTGCAAATTTTCTCTTTAGCAGAGCCAAATAACCTTGTTCTAGCTTTCTACTCGCAAAGTGCTTGTTTAAAGCAAATTGATTCCGGAAGATCTGTTGGCTCGACTTTGTTAATAATATTCTTTTCGTTTGTCATATTTTACTTGGTCCTCGGAGTTTGCACCAAGAAATTCCTGATGGGTGCAACTGGTGTGGAAGTTATACCCAATCTAGGATTTTGGACAGATCTGCCCAATCTTGTTAAGGATGGATGGGCATTTATGTTAAGCGGCTTCAAACTGCCAGCTCGTAGCGCTGGGCCTACACTGTCGCCTGACCCCAACAGCTATGACAGCATATAA
Protein
MDFRIKIYLFMSCLYCSIFPIYGEDVCLKKSACVCEFSNGTGIDLSSASKSTFFSAQTFEVAPATNAIELSTYYYHPCYDILLKPNSTTVNNTCTVPTAICRHREVLHAVDGTPDTYKKDLNVYDLIGNTDKSVFSAKGDAIVYTHGPSNTTVLLICSLSDGELQIFSLAEPNNLVLAFYSQSACLKQIDSGRSVGSTLLIIFFSFVIFYLVLGVCTKKFLMGATGVEVIPNLGFWTDLPNLVKDGWAFMLSGFKLPARSAGPTLSPDPNSYDSI
Summary
Uniprot
H9JFQ3
A0A194PNA6
A0A194QLZ9
A0A2A4K7U6
A0A1E1VYD8
A0A2W1BNZ1
+ More
A0A212EMZ9 A0A0L7KYS0 A0A0L7L493 B0WSP9 A0A182GVN3 A0A023EMS7 A0A1Q3FH18 A0A182F7S7 W5JL67 Q17M86 A0A182Q7A0 A0A2M4AV05 A0A2M4AV13 A0A2M4BXD2 A0A1Y1MWM1 U5ET17 A0A182VWQ5 A0A182LZR1 D6WXB8 A0A182RGY0 A0A2A3ENN5 A0A182YHK6 A0A182P990 R7U9Y1 A0A182SVS1 A0A182K193 A0A182N537 A0A182XHY5 A0A067R411 A0A154PBV1 A0A182HSW6 A0A182LEZ3 Q7PGI1 A0A3R7M703 A0A087ZNT4 A0A336LM85 A0A3M6U999 A0A195DG14 A0A1B6I6E9 A0A158NKQ7 A0A1B6GPE9 A0A0M9A0B9 A0A1S6QML4 A0A1B6DPL8 A0A210QNZ8 A0A1B6M2D3 A0A2J7QXB0 F4X3G6 A0A1S3HK55 A0A182UXM9 A0A2B4SXE3 A0A182UIJ7 H2Z8L3 A0A210QZW1 A0A2C9LWX2 K1PHQ2 A0A195BLU9 A0A2C9M702 A0A1S3I8H7 N6SZ24 U4TU87 A0A310S7F7 A0A195CKY9 A0A1S3HZV6 A0A210PP02 A0A1S3HJL9 A0A1S3I3H5 A0A3L8DBF8 A0A1W2WFE9 A0A210QP02 A0A1S3H195 A0A1J1J1R6 A0A2G8LMC3 A0A210R056 A0A1I8JAT6 C3ZP88
A0A212EMZ9 A0A0L7KYS0 A0A0L7L493 B0WSP9 A0A182GVN3 A0A023EMS7 A0A1Q3FH18 A0A182F7S7 W5JL67 Q17M86 A0A182Q7A0 A0A2M4AV05 A0A2M4AV13 A0A2M4BXD2 A0A1Y1MWM1 U5ET17 A0A182VWQ5 A0A182LZR1 D6WXB8 A0A182RGY0 A0A2A3ENN5 A0A182YHK6 A0A182P990 R7U9Y1 A0A182SVS1 A0A182K193 A0A182N537 A0A182XHY5 A0A067R411 A0A154PBV1 A0A182HSW6 A0A182LEZ3 Q7PGI1 A0A3R7M703 A0A087ZNT4 A0A336LM85 A0A3M6U999 A0A195DG14 A0A1B6I6E9 A0A158NKQ7 A0A1B6GPE9 A0A0M9A0B9 A0A1S6QML4 A0A1B6DPL8 A0A210QNZ8 A0A1B6M2D3 A0A2J7QXB0 F4X3G6 A0A1S3HK55 A0A182UXM9 A0A2B4SXE3 A0A182UIJ7 H2Z8L3 A0A210QZW1 A0A2C9LWX2 K1PHQ2 A0A195BLU9 A0A2C9M702 A0A1S3I8H7 N6SZ24 U4TU87 A0A310S7F7 A0A195CKY9 A0A1S3HZV6 A0A210PP02 A0A1S3HJL9 A0A1S3I3H5 A0A3L8DBF8 A0A1W2WFE9 A0A210QP02 A0A1S3H195 A0A1J1J1R6 A0A2G8LMC3 A0A210R056 A0A1I8JAT6 C3ZP88
Pubmed
EMBL
BABH01001859
KQ459604
KPI92600.1
KQ461196
KPJ06552.1
NWSH01000044
+ More
PCG80301.1 GDQN01011308 GDQN01000097 JAT79746.1 JAT90957.1 KZ149999 PZC75354.1 AGBW02013764 OWR42853.1 JTDY01004254 KOB68377.1 JTDY01003010 KOB70313.1 DS232075 EDS34008.1 JXUM01091415 KQ563913 KXJ73080.1 GAPW01003448 JAC10150.1 GFDL01008198 JAV26847.1 ADMH02001248 ETN63509.1 CH477208 EAT47758.1 AXCN02000175 GGFK01011221 MBW44542.1 GGFK01011289 MBW44610.1 GGFJ01008599 MBW57740.1 GEZM01021521 JAV88960.1 GANO01004571 JAB55300.1 AXCM01001329 KQ971361 EFA08006.1 KZ288204 PBC33310.1 AMQN01009627 KB305828 ELU00623.1 KK852714 KDR17899.1 KQ434857 KZC08894.1 APCN01000372 AAAB01008859 EAA44924.1 QCYY01001992 ROT73784.1 UFQT01000055 SSX19096.1 RCHS01001996 RMX50255.1 KQ980886 KYN11833.1 GECU01025207 JAS82499.1 ADTU01018966 GECZ01005536 JAS64233.1 KQ435784 KOX74577.1 KX830755 AQW35022.1 GEDC01009686 JAS27612.1 NEDP02002599 OWF50464.1 GEBQ01009900 JAT30077.1 NEVH01009382 PNF33225.1 GL888613 EGI58994.1 LSMT01000008 PFX33859.1 NEDP02001107 OWF54299.1 JH818149 EKC18449.1 KQ976439 KYM86704.1 APGK01058525 KB741291 ENN70493.1 KB630245 KB630279 KB631948 ERL83493.1 ERL83511.1 ERL87415.1 KQ773583 OAD52378.1 KQ977649 KYN00744.1 NEDP02005571 OWF38198.1 QOIP01000011 RLU17219.1 OWF50466.1 CVRI01000065 CRL05700.1 MRZV01000032 PIK61406.1 OWF54291.1 GG666656 EEN45585.1
PCG80301.1 GDQN01011308 GDQN01000097 JAT79746.1 JAT90957.1 KZ149999 PZC75354.1 AGBW02013764 OWR42853.1 JTDY01004254 KOB68377.1 JTDY01003010 KOB70313.1 DS232075 EDS34008.1 JXUM01091415 KQ563913 KXJ73080.1 GAPW01003448 JAC10150.1 GFDL01008198 JAV26847.1 ADMH02001248 ETN63509.1 CH477208 EAT47758.1 AXCN02000175 GGFK01011221 MBW44542.1 GGFK01011289 MBW44610.1 GGFJ01008599 MBW57740.1 GEZM01021521 JAV88960.1 GANO01004571 JAB55300.1 AXCM01001329 KQ971361 EFA08006.1 KZ288204 PBC33310.1 AMQN01009627 KB305828 ELU00623.1 KK852714 KDR17899.1 KQ434857 KZC08894.1 APCN01000372 AAAB01008859 EAA44924.1 QCYY01001992 ROT73784.1 UFQT01000055 SSX19096.1 RCHS01001996 RMX50255.1 KQ980886 KYN11833.1 GECU01025207 JAS82499.1 ADTU01018966 GECZ01005536 JAS64233.1 KQ435784 KOX74577.1 KX830755 AQW35022.1 GEDC01009686 JAS27612.1 NEDP02002599 OWF50464.1 GEBQ01009900 JAT30077.1 NEVH01009382 PNF33225.1 GL888613 EGI58994.1 LSMT01000008 PFX33859.1 NEDP02001107 OWF54299.1 JH818149 EKC18449.1 KQ976439 KYM86704.1 APGK01058525 KB741291 ENN70493.1 KB630245 KB630279 KB631948 ERL83493.1 ERL83511.1 ERL87415.1 KQ773583 OAD52378.1 KQ977649 KYN00744.1 NEDP02005571 OWF38198.1 QOIP01000011 RLU17219.1 OWF50466.1 CVRI01000065 CRL05700.1 MRZV01000032 PIK61406.1 OWF54291.1 GG666656 EEN45585.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000007151
UP000037510
+ More
UP000002320 UP000069940 UP000249989 UP000069272 UP000000673 UP000008820 UP000075886 UP000075920 UP000075883 UP000007266 UP000075900 UP000242457 UP000076408 UP000075885 UP000014760 UP000075901 UP000075881 UP000075884 UP000076407 UP000027135 UP000076502 UP000075840 UP000075882 UP000007062 UP000283509 UP000005203 UP000275408 UP000078492 UP000005205 UP000053105 UP000242188 UP000235965 UP000007755 UP000085678 UP000075903 UP000225706 UP000075902 UP000007875 UP000076420 UP000005408 UP000078540 UP000019118 UP000030742 UP000078542 UP000279307 UP000183832 UP000230750 UP000095280 UP000001554
UP000002320 UP000069940 UP000249989 UP000069272 UP000000673 UP000008820 UP000075886 UP000075920 UP000075883 UP000007266 UP000075900 UP000242457 UP000076408 UP000075885 UP000014760 UP000075901 UP000075881 UP000075884 UP000076407 UP000027135 UP000076502 UP000075840 UP000075882 UP000007062 UP000283509 UP000005203 UP000275408 UP000078492 UP000005205 UP000053105 UP000242188 UP000235965 UP000007755 UP000085678 UP000075903 UP000225706 UP000075902 UP000007875 UP000076420 UP000005408 UP000078540 UP000019118 UP000030742 UP000078542 UP000279307 UP000183832 UP000230750 UP000095280 UP000001554
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JFQ3
A0A194PNA6
A0A194QLZ9
A0A2A4K7U6
A0A1E1VYD8
A0A2W1BNZ1
+ More
A0A212EMZ9 A0A0L7KYS0 A0A0L7L493 B0WSP9 A0A182GVN3 A0A023EMS7 A0A1Q3FH18 A0A182F7S7 W5JL67 Q17M86 A0A182Q7A0 A0A2M4AV05 A0A2M4AV13 A0A2M4BXD2 A0A1Y1MWM1 U5ET17 A0A182VWQ5 A0A182LZR1 D6WXB8 A0A182RGY0 A0A2A3ENN5 A0A182YHK6 A0A182P990 R7U9Y1 A0A182SVS1 A0A182K193 A0A182N537 A0A182XHY5 A0A067R411 A0A154PBV1 A0A182HSW6 A0A182LEZ3 Q7PGI1 A0A3R7M703 A0A087ZNT4 A0A336LM85 A0A3M6U999 A0A195DG14 A0A1B6I6E9 A0A158NKQ7 A0A1B6GPE9 A0A0M9A0B9 A0A1S6QML4 A0A1B6DPL8 A0A210QNZ8 A0A1B6M2D3 A0A2J7QXB0 F4X3G6 A0A1S3HK55 A0A182UXM9 A0A2B4SXE3 A0A182UIJ7 H2Z8L3 A0A210QZW1 A0A2C9LWX2 K1PHQ2 A0A195BLU9 A0A2C9M702 A0A1S3I8H7 N6SZ24 U4TU87 A0A310S7F7 A0A195CKY9 A0A1S3HZV6 A0A210PP02 A0A1S3HJL9 A0A1S3I3H5 A0A3L8DBF8 A0A1W2WFE9 A0A210QP02 A0A1S3H195 A0A1J1J1R6 A0A2G8LMC3 A0A210R056 A0A1I8JAT6 C3ZP88
A0A212EMZ9 A0A0L7KYS0 A0A0L7L493 B0WSP9 A0A182GVN3 A0A023EMS7 A0A1Q3FH18 A0A182F7S7 W5JL67 Q17M86 A0A182Q7A0 A0A2M4AV05 A0A2M4AV13 A0A2M4BXD2 A0A1Y1MWM1 U5ET17 A0A182VWQ5 A0A182LZR1 D6WXB8 A0A182RGY0 A0A2A3ENN5 A0A182YHK6 A0A182P990 R7U9Y1 A0A182SVS1 A0A182K193 A0A182N537 A0A182XHY5 A0A067R411 A0A154PBV1 A0A182HSW6 A0A182LEZ3 Q7PGI1 A0A3R7M703 A0A087ZNT4 A0A336LM85 A0A3M6U999 A0A195DG14 A0A1B6I6E9 A0A158NKQ7 A0A1B6GPE9 A0A0M9A0B9 A0A1S6QML4 A0A1B6DPL8 A0A210QNZ8 A0A1B6M2D3 A0A2J7QXB0 F4X3G6 A0A1S3HK55 A0A182UXM9 A0A2B4SXE3 A0A182UIJ7 H2Z8L3 A0A210QZW1 A0A2C9LWX2 K1PHQ2 A0A195BLU9 A0A2C9M702 A0A1S3I8H7 N6SZ24 U4TU87 A0A310S7F7 A0A195CKY9 A0A1S3HZV6 A0A210PP02 A0A1S3HJL9 A0A1S3I3H5 A0A3L8DBF8 A0A1W2WFE9 A0A210QP02 A0A1S3H195 A0A1J1J1R6 A0A2G8LMC3 A0A210R056 A0A1I8JAT6 C3ZP88
Ontologies
Topology
SignalP
Position: 1 - 23,
Likelihood: 0.948962
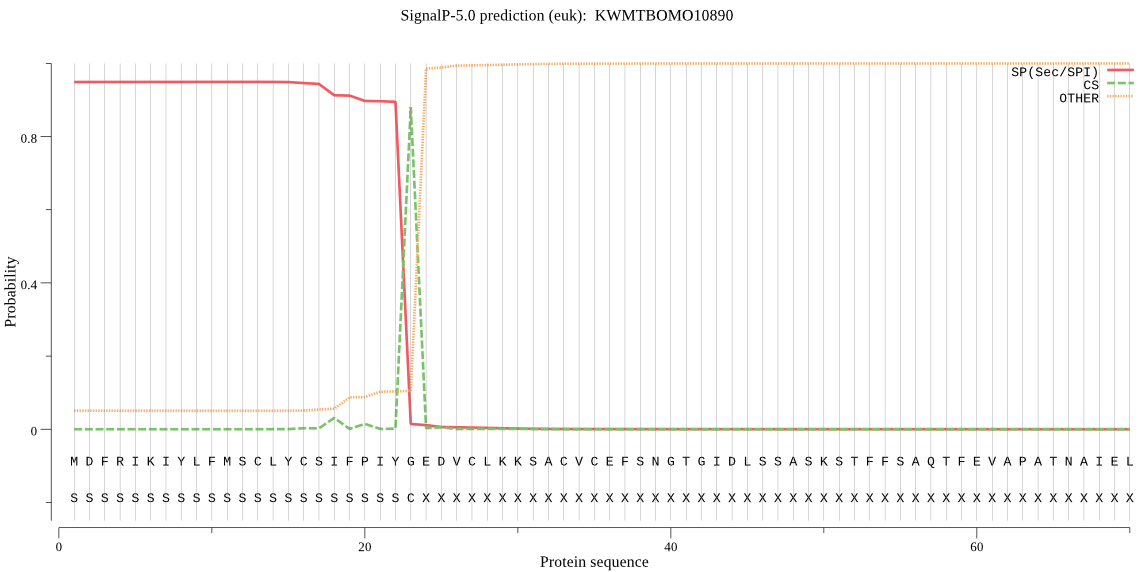
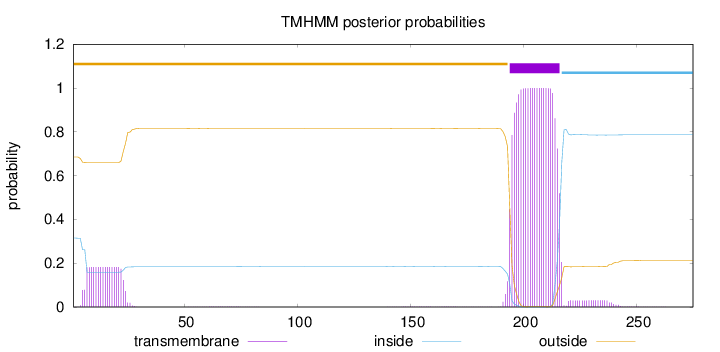
Length:
275
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
25.53228
Exp number, first 60 AAs:
3.32249
Total prob of N-in:
0.31507
outside
1 - 193
TMhelix
194 - 216
inside
217 - 275
Population Genetic Test Statistics
Pi
189.632746
Theta
178.344032
Tajima's D
-0.523778
CLR
34.470554
CSRT
0.24238788060597
Interpretation
Uncertain
